annealingnaning9官网
naning9官网 时间:2021-01-18 阅读:()
1of3ApplicationsinPlantSciences20153(8):1500040ApplicationsinPlantSciences20153(8):1500040;http://www.
bioone.
org/loi/apps2015Qinetal.
PublishedbytheBotanicalSocietyofAmerica.
ThisworkislicensedunderaCreativeCommonsAttributionLicense(CC-BY-NC-SA).
ApApplicatitionsonsininPlPlanttScienSciencescesIlexkaushueS.
Y.
Hu(Aquifoliaceae)isanevergreentreegrowingindenseforestsatelevationsbetween400and1000minsouthernChina(Chenetal.
,2008).
Thespeciesisoneoftheprimarysourcesofku-ding-cha(Haoetal.
,2013),ateausedintraditionalmedicinethathasbeenconsumedforthousandsofyearsinChina.
Modernmedicinalresearchhasdemonstratedthatku-ding-chahassignicantpharmacologicaleffects,in-cludingasanantidiabetesandantiobesitydrug,aswellasanantioxidant(Haoetal.
,2013).
Ilexkaushuelivesinafragmentedhabitatwithextremelysmallpopulationsizes,andwasincludedinaconservationprogramcarriedoutin2011bytheStateForestryAdministrationofChina(Chenetal.
,2014).
Furthermore,I.
kaushueisaneconomicallyimportantcropduetoitswide-spreadusefortea,andtherehasbeenrapiddevelopmentinitscultivationinsouthernChina(Guoetal.
,2005).
Evidencesupportsthatsmallnaturalpopulationsandmodernplantbreedingcanleadtoareductioninoverallgeneticdiver-sity(TanksleyandMcCouch,1997;Leimuetal.
,2006).
Lowlevelsofgeneticdiversityputwildpopulationsatriskandjeopardizethecontinuedabilitytoimprovecrops(Reifetal.
,2005).
Therefore,assessmentandpreservationofgeneticdiver-sityofI.
kaushueareimportantconcerns.
AlthoughZhangetal.
(2003)developedmolecularmarkers(RAPD)foruseinthestudyofI.
kaushue,furthergeneticdiversityresearchisnecessaryatthepopulationandspecieslevelstoassessandprotectitsgerm-plasmresources.
Assessmentandconservationofgeneticdiver-sityofaspeciesrequiresdevelopmentofefcientcodominantmicrosatellitemarkers.
Inthisstudy,16microsatellitelociforI.
kaushuewereisolatedandcharacterized,whichwillbeuse-fulforassessmentandconservationofgeneticdiversityofI.
kaushue.
METHODSANDRESULTSWesampled12I.
kaushuetreesinanaturalpopulation(BaishaCounty/HainanProvince,China[QS]:19°08′50.
38″N,109°16′14.
9″E)and10treesinacultivatedpopulation(DapuCounty/GuangdongProvince,China[DM]:24°16′40.
31″N,116°28′02.
83″E).
VoucherspecimensofeachpopulationweredepositedintheGuangxiInstituteofTraditionalMedicalandPharmaceuticalSciencesherbarium(GXMI;accessionnumbersIk-012-ZQWandIk-008-HYF,respectively;Appendix1).
GenomicDNA(gDNA)wasextractedfromsilicagel–driedleavesusingthecetyltrimethylammoniumbromide(CTAB)method(DoyleandDoyle,1987).
WemixedgDNAofallwild-collectedindividualstobeshotgunsequencedbySangonBiotech(Shanghai,China)using454GSFLXTitanium(RocheAppliedScience,Branford,Connecticut,USA).
The454sequencingtechniqueisdescribedindetailinMarguliesetal.
(2005).
Weobtained29,247readsrangingfrom32to691bpwithanaveragereadlengthof401bp,foratotalof11,736,223bases.
AllreadswerefurtherscreenedformicrosatellitemotifsimplementedintheprogramSSRHunter1.
3withthedefaultparameters(LiandWan,2005).
Atotalof1109sequencescontaining1104dinucleotide,259trinucleotide,andninetetranucleotiderepeatswereob-tained.
Ofthesesequences,thosecontainingatleastsixdinucleotideortrinu-cleotiderepeatsandsufcientlengthsateitherendoftherepeatmotifwerechosenforprimerdesignusingPrimerPremier5.
0(ClarkeandGorley,2001);atotalof631sequences,containing691dinucleotideand82trinucleotidere-peats,weresubjectedtoprimerdesign.
ThesettingsforPrimerPremierwereasfollows:(i)eachsearchrangeofsenseprimerandantisenseprimerwasateachendoftherepeatmotif;(ii)theprimerlengthwasbetween17and25bp;(iii)thePCRproductsizewasbetween100and400bplong;(iv)theannealingtemperatureofprimerswasbetween50°Cand64°C,andthedifferenceinannealingtemperaturebetweentheforwardandreverseprimerswas<4°C;(v)theGCcontentwasbetween40%and60%;(vi)therewasnotobvioushairpinstructurewithintheprimer;and(vii)otherparametersfollowedthedefaultsettingsof"High"stringencyinthesearchcriteria.
Atotalof78primerpairsweresuccessfullydesignedforatotalof99repeatsincluding65dinucleotide,1Manuscriptreceived3April2015;revisionaccepted6May2015.
ThisworkwassupportedbytheKeyProjectsintheNationalScienceandTechnologyPillarProgramduringtheTwelfthFive-YearPlanPeriod(2011BAI01B04)andbytheGuangxiScienticResearchandTechnologyDevelopmentProgram(14124002-3).
4Authorforcorrespondence:zqw_21@163.
comdoi:10.
3732/apps.
1500040PRIMERNOTEDEVELOPMENTOFMICROSATELLITEMARKERSINILEXKAUSHUE(AQUIFOLIACEAE),AMEDICINALPLANTSPECIES1LAN-FANGQIN2,XIN-CHENGQU2,GANGHU3,YUN-FENGHUANG2,ANDQI-WEIZHANG2,42GuangxiInstituteofTraditionalMedicalandPharmaceuticalSciences,Naning530022,People'sRepublicofChina;and3SchoolofEnvironmentandLifeSciences,GuangxiTeacher'sEducationUniversity,Naning530001,People'sRepublicofChinaPremiseofthestudy:MicrosatellitemarkersweredevelopedforIlexkaushue(Aquifoliaceae),amedicinalplantwithex-tremelysmallwildpopulationsthatexistsinfragmentedhabitats,toassessandprotectitsgeneticdiversity.
MethodsandResults:Using454GSFLXTitaniumsequencing,16microsatelliteprimersetswereisolatedandcharacterized.
Fifteenofthesemarkerswerepolymorphic.
Thenumberofallelesperlocusrangedfromonetonineacross22individualsfrombothcultivatedandwildpopulations.
Theobservedandexpectedheterozygosityinthesetwopopulationsrangedfrom0.
000to1.
000andfrom0.
000to0.
785,respectively.
Conclusions:ThesemarkerswillbeusefulinstudiesongeneticdiversityofI.
kaushue.
Keywords:Aquifoliaceae;geneticdiversity;Ilexkaushue;microsatellitemarker.
2of3ApplicationsinPlantSciences20153(8):1500040Qinetal.
—Ilexkaushuemicrosatellitesdoi:10.
3732/apps.
1500040http://www.
bioone.
org/loi/apps45s;andanalextensionstepat72°Cfor5min.
PCRproductswereresolvedon6%polyacrylamidedenaturinggelusinga10-bpor25-bpDNAladder(Invitrogen,Carlsbad,California,USA)asareferenceandwerevisualizedbysilverstaining.
Sixteenprimerpairsweresuccessfullyamplied;theseproductsexhibitedtheexpectedsizesandshowedclearlydenedbandingpatternswithamaximumoftwoallelesineachlocusperindividual.
Thenumberofallelesperlocus(A)andtheobservedandexpectedheterozygosity(HoandHe)ofthetwopopulationswereestimatedbyGenAlExversion6(PeakallandSmouse,2006).
Linkage18trinucleotide,and16compoundrepeats.
Theseprimersweretestedforpoly-morphismin22individualsfromthetwopopulations.
PCRreactionswereperformedina20-μLreactionvolumecontaining50–100ngofgDNA,0.
5μMofeachprimer,and10μLof2*TaqPCRMasterMix(0.
1U/μLTaqpolymerase,0.
5mMdNTPeach,20mMTris-HCl[pH8.
3],100mMKCl,and3mMMgCl2[TiangenBiotech,Beijing,China]).
PCRamplicationswereconductedunderthefollowingconditions:95°Cfor5min;followedby35cyclesat94°Cfor45s,attheannealingtemperatureforeachspecicprimer(optimizedforeachlocus,Table1)for45s,72°CforTABLE1.
Characteristicsof16microsatellitemarkersinIlexkaushue.
LocusPrimersequences(5′–3′)RepeatmotifTa(°C)Allelesizerange(bp)GenBankaccessionno.
KDC1F:CTTACTCCCTTTGGTGCTC(AG)1360181–191KP943496R:CTCTTTTAGTCATTTTGCCCKDC10F:GGCCCTCCTGTAATTTTTC(TA)758133–139KP943497R:GGTCGGTCCCATTCTTGTKDC11F:TCTCAGGGTGCCTAAATA(GA)756122–138KP868632R:AACTAAGGTGTTTAAGGTCCKDC12F:GTAGACGACAATAATGGCGG(TGG)660329–335KP868633R:CTCCACCGATTGCTACTATTGKDC16F:CGAGCGGAAAGCAGAAATC(GTG)660238KP943498R:AGCCGAGGCAGAGGTAAAGAKDC27F:GACAACCAAACACAGAAAAG(AG)857186–192KP868635R:CAAAAGGACCAGTAACCCKDC29F:GAGTGGTTTGTATGGTCTTGT(TG)7(GT)560203–207KP868636R:CAGTGGTTAGCCTTTGATTCKDC32F:AGGTGATAAAGGAGAGGTCG(AT)5(AG)760127–135KP868637R:CTCCCTCTCGTATACCACCTKDC41F:CACTAGTTGCATTGGTGCT(TTC)1058282–306KP868638R:TGTTTAATGAACCCACCTCKDC49F:CAACTAACCCTATGTGTC(AG)1453120–142KP868639R:TTGTTAGAAAATCCTCGKDC50F:GCATGGTCTTTTGAAAACGA(GA)1458272–286KP943499R:GGGACGGCATAGAACTGTAATKDC58F:AGAGGACAACGAAGATTAGG(CT)960346–350KP868641R:GAGAGGGTGGACTGAGAGATKDC61F:CATTCCACTGACACAACCG(GA)860238–244KP868642R:GAGCCTCCTCCTTCATTGTKDC62F:GTGTTGTTGATGGTGGGTT(GA)5(GA)759166–176KP868643R:ACGTTAGACCCACTCTCATCKDC63F:CGACATTTACAGTCTAGC(GT)856170–174KP868644R:CTCAACCTTTAACTCTCTCKDC66F:CCAACAAATCAATAGGGAC(GA)956142–156KP868645R:AACTTTTAAGAGCAGTGCCNote:Ta=annealingtemperaturewhenrunindividually.
TABLE2.
ResultsofinitialprimerscreeningintwopopulationsofIlexkaushue.
LocusQSpopulation(natural,N=12)DMpopulation(cultivated,N=10)ATAHoHePvalueAHoHePvalueKDC130.
6670.
6531.
00020.
3000.
3750.
4804KDC1030.
4170.
5690.
19320.
6000.
5000.
9764KDC1140.
6670.
5590.
60730.
3000.
5850.
0576KDC1230.
4170.
5690.
50420.
6000.
4801.
0003KDC161———1———1KDC2740.
7500.
7261.
0001———4KDC291———30.
8000.
6601.
0003KDC3240.
8330.
7120.
28620.
5000.
4951.
0005KDC4150.
8330.
7080.
53330.
8000.
6551.
0006KDC4960.
9170.
7640.
84650.
9000.
6750.
9539KDC5061.
0000.
7850.
79140.
7000.
6550.
8497KDC5830.
5830.
5691.
0001———3KDC6130.
5830.
5311.
00020.
5000.
4951.
0004KDC6250.
6670.
7260.
80920.
6000.
5001.
0005KDC6320.
3330.
5000.
28230.
6000.
6451.
0003KDC6640.
9170.
6910.
54730.
8000.
6650.
0246Note:A=numberofalleles;AT=totalnumberofalleles;He=expectedheterozygosity;Ho=observedheterozygosity;N=samplesizeforeachpopulation;Pvalue=testfordeviationfromHardy–Weinbergexpectations.
ApplicationsinPlantSciences20153(8):1500040Qinetal.
—Ilexkaushuemicrosatellitesdoi:10.
3732/apps.
15000403of3http://www.
bioone.
org/loi/appsdisequilibrium(LD)anddeviationfromHardy–Weinbergequilibrium(HWE)werecalculatedbyGENEPOPversion4.
2(RaymondandRousset,1995).
Acrossthecultivatedandwildpopulations,Avariedfromonetonine,andatotalof73alleleswerescoredin22individuals(Table2).
HoandHeinthenaturalandcultivatedpopulationsrangedfrom0.
000to1.
000andfrom0.
000to0.
785,respectively.
NopairsoflocishowedsignicantLD.
ThePvalueoftestsforHWErangedfrom0.
024to1.
000(Table2).
OnlylocusKDC66inpopulationDMsignicantlydeviatedfromHWE(P<0.
05),whichmayduetooverdominantselectionoradmixturefromdifferentresourcesgiventhehighlevelofheterozygosityforthislocus.
CONCLUSIONSAtotalof16nuclearmicrosatellitemarkersweredevelopedforI.
kaushue.
Fifteenofthesemarkersshowedvaryinglevelsofpolymorphismandonemarkerexhibitedmonomorphism.
TheselociwillbeusefulforassessmentandconservationofgeneticdiversityofI.
kaushue.
LITERATURECITEDCHEN,S.
K.
,H.
Y.
MA,Y.
X.
FENG,G.
BARRIERA,ANDP.
LOIZEAU.
2008.
Aquifoliaceae.
InZ.
Y.
WuandP.
H.
Raven[eds.
],FloraofChina,vol.
20,394.
SciencePress,Beijing,China,andMissouriBotanicalGardenPress,St.
Louis,Missouri,USA.
CHEN,Y.
K.
,X.
B.
YANG,Q.
YANG,D.
H.
LI,W.
X.
LONG,ANDW.
Q.
LUO.
2014.
FactorsaffectingthedistributionpatternofwildplantswithextremelysmallpopulationsinHainanIsland,China.
PLoSOne9:e97751.
CLARKE,K.
R.
,ANDR.
N.
GORLEY.
2001.
PRIMERv5:Usermanual/tutorial.
PRIMER-ELtd.
,Plymouth,UnitedKingdom.
DOYLE,J.
J.
,ANDJ.
L.
DOYLE.
1987.
ArapidDNAisolationprocedureforsmallquantitiesoffreshleaftissue.
PhytochemicalBulletin19:11–15.
GUO,L.
F.
,Q.
S.
JIANG,X.
G.
WANG,J.
X.
HE,ANDS.
Y.
JIANG.
2005.
Presentstatusandnon-pollutioncultivationtechniquesofIlexkudingchainGuangxi.
Guihaia25:366–371.
HAO,D.
C.
,X.
J.
GU,P.
J.
XIAO,Z.
G.
LIANG,L.
J.
XU,ANDY.
PENG.
2013.
ResearchprogressinthephytochemistryandbiologyofIlexpharma-ceuticalresources.
ActaPharmaceuticaSinica.
B3:8–19.
LEIMU,R.
,P.
MUTIKANINEN,J.
KORICHEVA,ANDM.
FISCHER.
2006.
Howgeneralarepositiverelationshipsbetweenplantpopulationsize,t-nessandgeneticvariationJournalofEcology94:942–952.
LI,Q.
,ANDJ.
M.
WAN.
2005.
SSRHunter:Developmentofalocalsearch-ingsoftwareforSSRsites.
Hereditas27:808–810.
MARGULIES,M.
,M.
EGHOLM,W.
E.
ALTMAN,S.
ATTIYA,J.
S.
BADER,L.
A.
BEMBEN,J.
BERKA,ETAL.
2005.
Genomesequencinginmicrofabri-catedhigh-densitypicolitrereactors.
Nature437:376–380.
PEAKALL,R.
,ANDP.
E.
SMOUSE.
2006.
GenAlEx6:GeneticanalysisinExcel.
Populationgeneticsoftwareforteachingandresearch.
MolecularEcologyNotes6:288–295.
RAYMOND,M.
,ANDF.
ROUSSET.
1995.
GENEPOP(version1.
2):Populationgeneticsoftwareforexacttestsandecumenicism.
JournalofHeredity86:248–249.
REIF,J.
C.
,P.
ZHANG,S.
DREISIGACKER,M.
L.
WARBURTON,M.
VANGINKEL,D.
HOISINGTON,M.
BOHN,ANDA.
E.
MELCHINGER.
2005.
Wheatge-neticdiversitytrendsduringdomesticationandbreeding.
TheoreticalandAppliedGenetics110:859–864.
TANKSLEY,S.
D.
,ANDS.
R.
MCCOUCH.
1997.
Seedbanksandmolecu-larmaps:Unlockinggeneticpotentialfromthewild.
Science277:1063–1066.
ZHANG,F.
Q.
,L.
X.
XU,P.
ZHOU,G.
M.
LIU,A.
P.
GUO,ANDQ.
T.
QIU.
2003.
TheinuentialfactorsofRAPDinIlexkudingchaandtheop-timizationoftheexperimentalconditions.
ActaBotanicaYunnanica25:347–353.
APPENDIX1.
VoucherinformationforIlexkaushueusedinthisstudy.
PopulationVoucherspecimenaccessionno.
aCollectionlocalitybGeographiccoordinatesNQSIk-012-ZQWQingsongTownship,BaishaCounty,HainanProvince19°08′50.
38″N,109°16′14.
91″E12DMIk-008-HYFDamaTown,DapuCounty,GuangdongProvince24°16′40.
31″N,116°28′02.
83″E10Note:N=numberofindividuals.
aVouchersdepositedintheGuangxiInstituteofTraditionalMedicalandPharmaceuticalSciencesherbarium.
ZQW=Qi-WeiZhang,collector;HYF=Yun-FengHuang,collector.
bLocalityandChineseprovince.
bioone.
org/loi/apps2015Qinetal.
PublishedbytheBotanicalSocietyofAmerica.
ThisworkislicensedunderaCreativeCommonsAttributionLicense(CC-BY-NC-SA).
ApApplicatitionsonsininPlPlanttScienSciencescesIlexkaushueS.
Y.
Hu(Aquifoliaceae)isanevergreentreegrowingindenseforestsatelevationsbetween400and1000minsouthernChina(Chenetal.
,2008).
Thespeciesisoneoftheprimarysourcesofku-ding-cha(Haoetal.
,2013),ateausedintraditionalmedicinethathasbeenconsumedforthousandsofyearsinChina.
Modernmedicinalresearchhasdemonstratedthatku-ding-chahassignicantpharmacologicaleffects,in-cludingasanantidiabetesandantiobesitydrug,aswellasanantioxidant(Haoetal.
,2013).
Ilexkaushuelivesinafragmentedhabitatwithextremelysmallpopulationsizes,andwasincludedinaconservationprogramcarriedoutin2011bytheStateForestryAdministrationofChina(Chenetal.
,2014).
Furthermore,I.
kaushueisaneconomicallyimportantcropduetoitswide-spreadusefortea,andtherehasbeenrapiddevelopmentinitscultivationinsouthernChina(Guoetal.
,2005).
Evidencesupportsthatsmallnaturalpopulationsandmodernplantbreedingcanleadtoareductioninoverallgeneticdiver-sity(TanksleyandMcCouch,1997;Leimuetal.
,2006).
Lowlevelsofgeneticdiversityputwildpopulationsatriskandjeopardizethecontinuedabilitytoimprovecrops(Reifetal.
,2005).
Therefore,assessmentandpreservationofgeneticdiver-sityofI.
kaushueareimportantconcerns.
AlthoughZhangetal.
(2003)developedmolecularmarkers(RAPD)foruseinthestudyofI.
kaushue,furthergeneticdiversityresearchisnecessaryatthepopulationandspecieslevelstoassessandprotectitsgerm-plasmresources.
Assessmentandconservationofgeneticdiver-sityofaspeciesrequiresdevelopmentofefcientcodominantmicrosatellitemarkers.
Inthisstudy,16microsatellitelociforI.
kaushuewereisolatedandcharacterized,whichwillbeuse-fulforassessmentandconservationofgeneticdiversityofI.
kaushue.
METHODSANDRESULTSWesampled12I.
kaushuetreesinanaturalpopulation(BaishaCounty/HainanProvince,China[QS]:19°08′50.
38″N,109°16′14.
9″E)and10treesinacultivatedpopulation(DapuCounty/GuangdongProvince,China[DM]:24°16′40.
31″N,116°28′02.
83″E).
VoucherspecimensofeachpopulationweredepositedintheGuangxiInstituteofTraditionalMedicalandPharmaceuticalSciencesherbarium(GXMI;accessionnumbersIk-012-ZQWandIk-008-HYF,respectively;Appendix1).
GenomicDNA(gDNA)wasextractedfromsilicagel–driedleavesusingthecetyltrimethylammoniumbromide(CTAB)method(DoyleandDoyle,1987).
WemixedgDNAofallwild-collectedindividualstobeshotgunsequencedbySangonBiotech(Shanghai,China)using454GSFLXTitanium(RocheAppliedScience,Branford,Connecticut,USA).
The454sequencingtechniqueisdescribedindetailinMarguliesetal.
(2005).
Weobtained29,247readsrangingfrom32to691bpwithanaveragereadlengthof401bp,foratotalof11,736,223bases.
AllreadswerefurtherscreenedformicrosatellitemotifsimplementedintheprogramSSRHunter1.
3withthedefaultparameters(LiandWan,2005).
Atotalof1109sequencescontaining1104dinucleotide,259trinucleotide,andninetetranucleotiderepeatswereob-tained.
Ofthesesequences,thosecontainingatleastsixdinucleotideortrinu-cleotiderepeatsandsufcientlengthsateitherendoftherepeatmotifwerechosenforprimerdesignusingPrimerPremier5.
0(ClarkeandGorley,2001);atotalof631sequences,containing691dinucleotideand82trinucleotidere-peats,weresubjectedtoprimerdesign.
ThesettingsforPrimerPremierwereasfollows:(i)eachsearchrangeofsenseprimerandantisenseprimerwasateachendoftherepeatmotif;(ii)theprimerlengthwasbetween17and25bp;(iii)thePCRproductsizewasbetween100and400bplong;(iv)theannealingtemperatureofprimerswasbetween50°Cand64°C,andthedifferenceinannealingtemperaturebetweentheforwardandreverseprimerswas<4°C;(v)theGCcontentwasbetween40%and60%;(vi)therewasnotobvioushairpinstructurewithintheprimer;and(vii)otherparametersfollowedthedefaultsettingsof"High"stringencyinthesearchcriteria.
Atotalof78primerpairsweresuccessfullydesignedforatotalof99repeatsincluding65dinucleotide,1Manuscriptreceived3April2015;revisionaccepted6May2015.
ThisworkwassupportedbytheKeyProjectsintheNationalScienceandTechnologyPillarProgramduringtheTwelfthFive-YearPlanPeriod(2011BAI01B04)andbytheGuangxiScienticResearchandTechnologyDevelopmentProgram(14124002-3).
4Authorforcorrespondence:zqw_21@163.
comdoi:10.
3732/apps.
1500040PRIMERNOTEDEVELOPMENTOFMICROSATELLITEMARKERSINILEXKAUSHUE(AQUIFOLIACEAE),AMEDICINALPLANTSPECIES1LAN-FANGQIN2,XIN-CHENGQU2,GANGHU3,YUN-FENGHUANG2,ANDQI-WEIZHANG2,42GuangxiInstituteofTraditionalMedicalandPharmaceuticalSciences,Naning530022,People'sRepublicofChina;and3SchoolofEnvironmentandLifeSciences,GuangxiTeacher'sEducationUniversity,Naning530001,People'sRepublicofChinaPremiseofthestudy:MicrosatellitemarkersweredevelopedforIlexkaushue(Aquifoliaceae),amedicinalplantwithex-tremelysmallwildpopulationsthatexistsinfragmentedhabitats,toassessandprotectitsgeneticdiversity.
MethodsandResults:Using454GSFLXTitaniumsequencing,16microsatelliteprimersetswereisolatedandcharacterized.
Fifteenofthesemarkerswerepolymorphic.
Thenumberofallelesperlocusrangedfromonetonineacross22individualsfrombothcultivatedandwildpopulations.
Theobservedandexpectedheterozygosityinthesetwopopulationsrangedfrom0.
000to1.
000andfrom0.
000to0.
785,respectively.
Conclusions:ThesemarkerswillbeusefulinstudiesongeneticdiversityofI.
kaushue.
Keywords:Aquifoliaceae;geneticdiversity;Ilexkaushue;microsatellitemarker.
2of3ApplicationsinPlantSciences20153(8):1500040Qinetal.
—Ilexkaushuemicrosatellitesdoi:10.
3732/apps.
1500040http://www.
bioone.
org/loi/apps45s;andanalextensionstepat72°Cfor5min.
PCRproductswereresolvedon6%polyacrylamidedenaturinggelusinga10-bpor25-bpDNAladder(Invitrogen,Carlsbad,California,USA)asareferenceandwerevisualizedbysilverstaining.
Sixteenprimerpairsweresuccessfullyamplied;theseproductsexhibitedtheexpectedsizesandshowedclearlydenedbandingpatternswithamaximumoftwoallelesineachlocusperindividual.
Thenumberofallelesperlocus(A)andtheobservedandexpectedheterozygosity(HoandHe)ofthetwopopulationswereestimatedbyGenAlExversion6(PeakallandSmouse,2006).
Linkage18trinucleotide,and16compoundrepeats.
Theseprimersweretestedforpoly-morphismin22individualsfromthetwopopulations.
PCRreactionswereperformedina20-μLreactionvolumecontaining50–100ngofgDNA,0.
5μMofeachprimer,and10μLof2*TaqPCRMasterMix(0.
1U/μLTaqpolymerase,0.
5mMdNTPeach,20mMTris-HCl[pH8.
3],100mMKCl,and3mMMgCl2[TiangenBiotech,Beijing,China]).
PCRamplicationswereconductedunderthefollowingconditions:95°Cfor5min;followedby35cyclesat94°Cfor45s,attheannealingtemperatureforeachspecicprimer(optimizedforeachlocus,Table1)for45s,72°CforTABLE1.
Characteristicsof16microsatellitemarkersinIlexkaushue.
LocusPrimersequences(5′–3′)RepeatmotifTa(°C)Allelesizerange(bp)GenBankaccessionno.
KDC1F:CTTACTCCCTTTGGTGCTC(AG)1360181–191KP943496R:CTCTTTTAGTCATTTTGCCCKDC10F:GGCCCTCCTGTAATTTTTC(TA)758133–139KP943497R:GGTCGGTCCCATTCTTGTKDC11F:TCTCAGGGTGCCTAAATA(GA)756122–138KP868632R:AACTAAGGTGTTTAAGGTCCKDC12F:GTAGACGACAATAATGGCGG(TGG)660329–335KP868633R:CTCCACCGATTGCTACTATTGKDC16F:CGAGCGGAAAGCAGAAATC(GTG)660238KP943498R:AGCCGAGGCAGAGGTAAAGAKDC27F:GACAACCAAACACAGAAAAG(AG)857186–192KP868635R:CAAAAGGACCAGTAACCCKDC29F:GAGTGGTTTGTATGGTCTTGT(TG)7(GT)560203–207KP868636R:CAGTGGTTAGCCTTTGATTCKDC32F:AGGTGATAAAGGAGAGGTCG(AT)5(AG)760127–135KP868637R:CTCCCTCTCGTATACCACCTKDC41F:CACTAGTTGCATTGGTGCT(TTC)1058282–306KP868638R:TGTTTAATGAACCCACCTCKDC49F:CAACTAACCCTATGTGTC(AG)1453120–142KP868639R:TTGTTAGAAAATCCTCGKDC50F:GCATGGTCTTTTGAAAACGA(GA)1458272–286KP943499R:GGGACGGCATAGAACTGTAATKDC58F:AGAGGACAACGAAGATTAGG(CT)960346–350KP868641R:GAGAGGGTGGACTGAGAGATKDC61F:CATTCCACTGACACAACCG(GA)860238–244KP868642R:GAGCCTCCTCCTTCATTGTKDC62F:GTGTTGTTGATGGTGGGTT(GA)5(GA)759166–176KP868643R:ACGTTAGACCCACTCTCATCKDC63F:CGACATTTACAGTCTAGC(GT)856170–174KP868644R:CTCAACCTTTAACTCTCTCKDC66F:CCAACAAATCAATAGGGAC(GA)956142–156KP868645R:AACTTTTAAGAGCAGTGCCNote:Ta=annealingtemperaturewhenrunindividually.
TABLE2.
ResultsofinitialprimerscreeningintwopopulationsofIlexkaushue.
LocusQSpopulation(natural,N=12)DMpopulation(cultivated,N=10)ATAHoHePvalueAHoHePvalueKDC130.
6670.
6531.
00020.
3000.
3750.
4804KDC1030.
4170.
5690.
19320.
6000.
5000.
9764KDC1140.
6670.
5590.
60730.
3000.
5850.
0576KDC1230.
4170.
5690.
50420.
6000.
4801.
0003KDC161———1———1KDC2740.
7500.
7261.
0001———4KDC291———30.
8000.
6601.
0003KDC3240.
8330.
7120.
28620.
5000.
4951.
0005KDC4150.
8330.
7080.
53330.
8000.
6551.
0006KDC4960.
9170.
7640.
84650.
9000.
6750.
9539KDC5061.
0000.
7850.
79140.
7000.
6550.
8497KDC5830.
5830.
5691.
0001———3KDC6130.
5830.
5311.
00020.
5000.
4951.
0004KDC6250.
6670.
7260.
80920.
6000.
5001.
0005KDC6320.
3330.
5000.
28230.
6000.
6451.
0003KDC6640.
9170.
6910.
54730.
8000.
6650.
0246Note:A=numberofalleles;AT=totalnumberofalleles;He=expectedheterozygosity;Ho=observedheterozygosity;N=samplesizeforeachpopulation;Pvalue=testfordeviationfromHardy–Weinbergexpectations.
ApplicationsinPlantSciences20153(8):1500040Qinetal.
—Ilexkaushuemicrosatellitesdoi:10.
3732/apps.
15000403of3http://www.
bioone.
org/loi/appsdisequilibrium(LD)anddeviationfromHardy–Weinbergequilibrium(HWE)werecalculatedbyGENEPOPversion4.
2(RaymondandRousset,1995).
Acrossthecultivatedandwildpopulations,Avariedfromonetonine,andatotalof73alleleswerescoredin22individuals(Table2).
HoandHeinthenaturalandcultivatedpopulationsrangedfrom0.
000to1.
000andfrom0.
000to0.
785,respectively.
NopairsoflocishowedsignicantLD.
ThePvalueoftestsforHWErangedfrom0.
024to1.
000(Table2).
OnlylocusKDC66inpopulationDMsignicantlydeviatedfromHWE(P<0.
05),whichmayduetooverdominantselectionoradmixturefromdifferentresourcesgiventhehighlevelofheterozygosityforthislocus.
CONCLUSIONSAtotalof16nuclearmicrosatellitemarkersweredevelopedforI.
kaushue.
Fifteenofthesemarkersshowedvaryinglevelsofpolymorphismandonemarkerexhibitedmonomorphism.
TheselociwillbeusefulforassessmentandconservationofgeneticdiversityofI.
kaushue.
LITERATURECITEDCHEN,S.
K.
,H.
Y.
MA,Y.
X.
FENG,G.
BARRIERA,ANDP.
LOIZEAU.
2008.
Aquifoliaceae.
InZ.
Y.
WuandP.
H.
Raven[eds.
],FloraofChina,vol.
20,394.
SciencePress,Beijing,China,andMissouriBotanicalGardenPress,St.
Louis,Missouri,USA.
CHEN,Y.
K.
,X.
B.
YANG,Q.
YANG,D.
H.
LI,W.
X.
LONG,ANDW.
Q.
LUO.
2014.
FactorsaffectingthedistributionpatternofwildplantswithextremelysmallpopulationsinHainanIsland,China.
PLoSOne9:e97751.
CLARKE,K.
R.
,ANDR.
N.
GORLEY.
2001.
PRIMERv5:Usermanual/tutorial.
PRIMER-ELtd.
,Plymouth,UnitedKingdom.
DOYLE,J.
J.
,ANDJ.
L.
DOYLE.
1987.
ArapidDNAisolationprocedureforsmallquantitiesoffreshleaftissue.
PhytochemicalBulletin19:11–15.
GUO,L.
F.
,Q.
S.
JIANG,X.
G.
WANG,J.
X.
HE,ANDS.
Y.
JIANG.
2005.
Presentstatusandnon-pollutioncultivationtechniquesofIlexkudingchainGuangxi.
Guihaia25:366–371.
HAO,D.
C.
,X.
J.
GU,P.
J.
XIAO,Z.
G.
LIANG,L.
J.
XU,ANDY.
PENG.
2013.
ResearchprogressinthephytochemistryandbiologyofIlexpharma-ceuticalresources.
ActaPharmaceuticaSinica.
B3:8–19.
LEIMU,R.
,P.
MUTIKANINEN,J.
KORICHEVA,ANDM.
FISCHER.
2006.
Howgeneralarepositiverelationshipsbetweenplantpopulationsize,t-nessandgeneticvariationJournalofEcology94:942–952.
LI,Q.
,ANDJ.
M.
WAN.
2005.
SSRHunter:Developmentofalocalsearch-ingsoftwareforSSRsites.
Hereditas27:808–810.
MARGULIES,M.
,M.
EGHOLM,W.
E.
ALTMAN,S.
ATTIYA,J.
S.
BADER,L.
A.
BEMBEN,J.
BERKA,ETAL.
2005.
Genomesequencinginmicrofabri-catedhigh-densitypicolitrereactors.
Nature437:376–380.
PEAKALL,R.
,ANDP.
E.
SMOUSE.
2006.
GenAlEx6:GeneticanalysisinExcel.
Populationgeneticsoftwareforteachingandresearch.
MolecularEcologyNotes6:288–295.
RAYMOND,M.
,ANDF.
ROUSSET.
1995.
GENEPOP(version1.
2):Populationgeneticsoftwareforexacttestsandecumenicism.
JournalofHeredity86:248–249.
REIF,J.
C.
,P.
ZHANG,S.
DREISIGACKER,M.
L.
WARBURTON,M.
VANGINKEL,D.
HOISINGTON,M.
BOHN,ANDA.
E.
MELCHINGER.
2005.
Wheatge-neticdiversitytrendsduringdomesticationandbreeding.
TheoreticalandAppliedGenetics110:859–864.
TANKSLEY,S.
D.
,ANDS.
R.
MCCOUCH.
1997.
Seedbanksandmolecu-larmaps:Unlockinggeneticpotentialfromthewild.
Science277:1063–1066.
ZHANG,F.
Q.
,L.
X.
XU,P.
ZHOU,G.
M.
LIU,A.
P.
GUO,ANDQ.
T.
QIU.
2003.
TheinuentialfactorsofRAPDinIlexkudingchaandtheop-timizationoftheexperimentalconditions.
ActaBotanicaYunnanica25:347–353.
APPENDIX1.
VoucherinformationforIlexkaushueusedinthisstudy.
PopulationVoucherspecimenaccessionno.
aCollectionlocalitybGeographiccoordinatesNQSIk-012-ZQWQingsongTownship,BaishaCounty,HainanProvince19°08′50.
38″N,109°16′14.
91″E12DMIk-008-HYFDamaTown,DapuCounty,GuangdongProvince24°16′40.
31″N,116°28′02.
83″E10Note:N=numberofindividuals.
aVouchersdepositedintheGuangxiInstituteofTraditionalMedicalandPharmaceuticalSciencesherbarium.
ZQW=Qi-WeiZhang,collector;HYF=Yun-FengHuang,collector.
bLocalityandChineseprovince.
- annealingnaning9官网相关文档
- fullynaning9官网
- MEMS测试分析平台
- https://elearning.shisu.edu.cn/
- ingnaning9官网
- eingreiftnaning9官网
- conceptnaning9官网
HostYun(月18元),CN2直连香港大带宽VPS 50M带宽起
对于如今的云服务商的竞争着实很激烈,我们可以看到国内国外服务商的各种内卷,使得我们很多个人服务商压力还是比较大的。我们看到这几年的服务商变动还是比较大的,很多新服务商坚持不超过三个月,有的是多个品牌同步进行然后分别的跑路赚一波走人。对于我们用户来说,便宜的服务商固然可以试试,但是如果是不确定的,建议月付或者主力业务尽量的还是注意备份。HostYun 最近几个月还是比较活跃的,在前面也有多次介绍到商...
美国G口/香港CTG/美国T级超防云/物理机/CDN大促销 1核 1G 24元/月
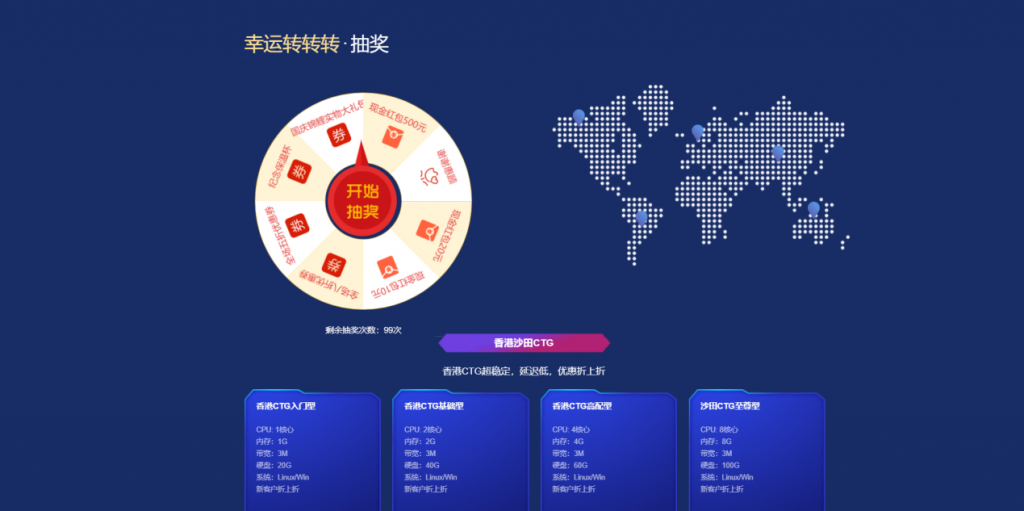
[六一云迎国庆]转盘活动实物礼品美国G口/香港CTG/美国T级超防云/物理机/CDN大促销六一云 成立于2018年,归属于西安六一网络科技有限公司,是一家国内正规持有IDC ISP CDN IRCS电信经营许可证书的老牌商家。大陆持证公司受大陆各部门监管不好用支持退款退现,再也不怕被割韭菜了!主要业务有:国内高防云,美国高防云,美国cera大带宽,香港CTG,香港沙田CN2,海外站群服务,物理机,...
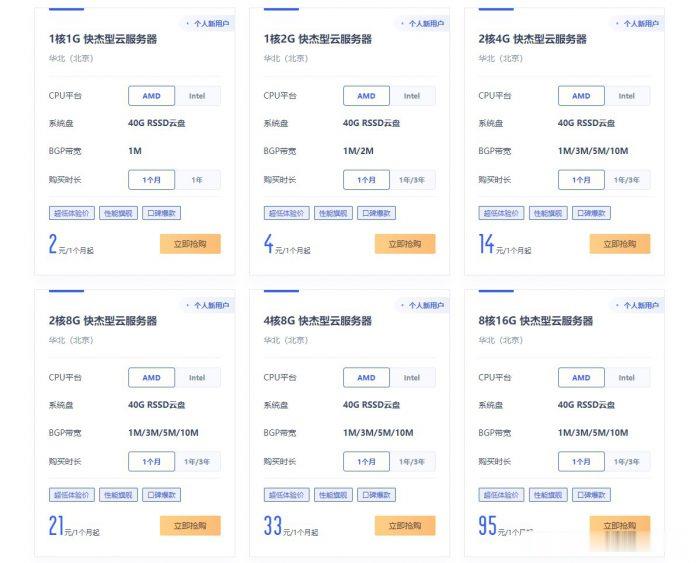
ucloud国内云服务器2元/月起;香港云服务器4元/首月;台湾云服务器3元/首月
ucloud云服务器怎么样?ucloud为了扩大云服务器市场份额,给出了超低价云服务器的促销活动,活动仍然是此前的Ucloud全球大促活动页面。目前,ucloud国内云服务器2元/月起;香港云服务器4元/首月;台湾云服务器3元/首月。相当于2-4元就可以试用国内、中国香港、中国台湾这三个地域的云服务器1个月了。ucloud全球大促仅限新用户,国内云服务器个人用户低至56元/年起,香港云服务器也仅8...
naning9官网为你推荐
-
国内域名注册预留的国内(cn)域名申请方法域名注册网网站域名申请,在那备案?已备案域名查询如何查询已备案的域名是否在万网备案的?vps主机什么是vps主机免费vps服务器免费VPS服务器。和免费的好用虚拟主机成都虚拟空间空间服务商那个好深圳网站空间深圳网站设计 哪家好一些?韩国虚拟主机香港和韩国的虚拟主机哪个比较好?虚拟主机管理系统如何用win虚拟主机管理系统搭建虚拟主机软件常见的虚拟机软件有哪几种?