hnRNPswww.bu622.com
www.bu622.com 时间:2021-05-08 阅读:()
RESEARCHOpenAccessProteomicprofileofpre-B2lymphoblastsfromchildrenwithacutelymphoblasticleukemia(ALL)inrelationwiththetranslocation(12;21)OdileCosta1*,PascaleSchneider1,2*,LaurentCoquet3,4,PhilippeChan3,DominiquePenther5,ElisabethLegrand1,ThierryJouenne3,4,MarcVasse1andJean-PierreVannier1,2AbstractBackground:Untilnow,themajorprognosticfactorsforpediatricacutelymphoblasticleukemia(ALL),age,whitebloodcellcountandchromosomalalterationsareinitiallytakenintoaccountfortheriskstratificationofpatients.
InthelightofproteinmarkerstudiestoclassifysubtypesofAcuteMyeloblasticLeukemiaefficiently,wehavecomparedthelymphoblastesproteomeinChildhoodALLinaccordancewiththepresenceoft(12;21),indicatorofgoodprognosis,usually.
Methods:Proteinexpressioninpre-B2lymphoblasticcells,collectedfromresidualbonemarrowcellsafterdiagnosticprocedures,wasanalyzedusingtwodimensionalgelelectrophoresisprotocol.
Proteinspotswhoseaveragenormalizedvolumeswerestatisticallydifferentinthetwopatientsgroups(n=13;studentttestp48)(seeOnlineAdditionalfile1:FigureS3)wereofgreatinterestintermsofleukemogenesissincein-volvedinmechanismsrelatedtoeitherthecellcycleregu-lation,orapoptosis,orenergeticmetabolism(i.
e.
,43%,29%or28%ofthem,respectively)(Figure2B).
PrincipalCostaetal.
ClinicalProteomics2014,11:31Page2of12http://www.
clinicalproteomicsjournal.
com/content/11/1/31Table1Clinicalandbiologicalstatusofchildhoodpre-B2ALLpatientsGroupofALLpatientsPatientsIDAgeatdiagnosis(Months)Peripheralbloodlymphocytes(Giga/L)KaryotypeFusiontranscriptsOutcomeWitht(12;21)BACYO2448,7t(12;21)ETV6/RUNX1CR1GUEKE11842,2t(12;21)ETV6/RUNX1CR1LECRO765,4t(12;21)ETV6/RUNX1CR1MARLE42166t(12;21)ETV6/RUNX1CR1PELAD5020t(12;21)ETV6/RUNX1Dead*RIFEM76216t(12;21)ETV6/RUNX1CR1OtherkaryotypeabnormalitiesBOILE182,8dic(9;20)NACR1CREBA18011,6mono21NACR1DESNA1315,5rear9pNACR1JUGEL348,5HyperploidyNACR1LEMAL4856t(9;22)BCL/ABL1CR1MORLU171171t(9;22)BCL/ABL1Dead*ROSLO576,1HyperploidyNACR1TheboldnumbersindicatesvalueshigherthethresholdofFRALLE2000protocol.
CR1=Firstcompleteremission;dic=dicentric;ID=Identification;mono=monosomy;NA=Notapplicable;rear=Rearrangement.
*=DirectlyrelatedtotherecurrenceofALL.
Figure1Differentialexpressionofproteinsinbonemarrowlymphoblasticcellsfromchildhoodpre-BALL.
–A–Down:Two-DEgelimageoflymphoblasticcellsproteinsissuingfrombonemarrow.
NumbersrefertothetopelevenproteinswhichhavebeencharacterizedandlistedinTable1b.
Up:Magnificationviewofinterestingzone–B–3Dmontageofthethreetopdifferentiallyexpressedspotproteins(i.
e.
,2-DEBatch2768,3185and4083onrank1,2and3,respectively).
Only2individualspotimagesineachgroup(1and2=Pre-B2ALLwitht(12;21),1′and2′=Pre-B2ALLwithotherkaryotypeabnormalities)areshown.
Costaetal.
ClinicalProteomics2014,11:31Page3of12http://www.
clinicalproteomicsjournal.
com/content/11/1/31Table2Theclassificationofdifferentiallyexpressedproteinsinpre-B2lymphoblasticcellsfromchildrenwithALLSpots:statisticrank(2-DEbatch)Proteinnames[Homosapiens]NCBIanduniprotIDnumbersFoldchanges/ALLt(12;21)versusothers(averagenormalizedvolume+/SD)105GroupwiththeupperaveragevolumeStudent-ttestPower(afterqvalue)1(2768)CNN2gi/49456619/Q994391.
9/(38.
59+/8.
1vs20.
76+/4)Pre-B2,t(12;21)p≤0.
0010.
9982(3185)CDSαgi/32307132/Q9Y697-12.
2/(9.
5+/3.
8vs20.
98+/5.
2)OtherPre-B2p≤0.
0010.
9853(4083)PITPβgi/6912594/P487392/(12.
8+/3.
1vs6.
28+/2.
21)Pre-B2,t(12;21)p≤0.
0010.
9834(2457)hnRNP-E1gi/460771/Q153651.
4/(29.
87+/4.
69vs41.
57+/4.
88)OtherPre-B2p≤0.
0050.
976BUB3αgi/4757880/O436845(4069)PDH-E1gi/149242791/P13804-11.
5/(15.
16+/4.
33vs23.
29+/3.
12)OtherPre-B2p≤0.
0050.
9556(2090)MAT2βi1gi/11034825/Q9NZL91.
5/(21.
94+/3.
89vs15.
06+/2.
91)Pre-B2,t(12;21)p≤0.
0050.
9157(2800)PSMB2gi/4506195/B7Z4781.
7/(9.
48+/2.
49vs5.
69+/1.
63)Pre-B2,t(12;21)p≤0.
010.
8788(4081)CECR5gi/14861834/Q9BXW71.
3/(28.
51+/4.
48vs37.
3+/4.
81)OtherPre-B2p≤0.
010.
867BUB3αgi/4757880/O436849(3121)CK2αgi/4503095/P684001.
5/(13.
55+/2.
36vs20.
31+/4.
9)OtherPre-B2p≤0.
010.
864SEPT9i3gi/668381/Q9UHD8-310(2056)HnRNPA2gi/4504447/P22626-22/(18.
28+/6.
56vs9.
26+/4.
77)Pre-B2,t(12;21)p≤0.
010.
85411(3120)IVADgi/3212539/P264401.
5/(14.
98+/3.
44vs22.
64+/5.
34)OtherPreB2p≤0.
010.
835FBAgi/312137/P05062PSMB6gi/1526426/P6233312(2656)OTUB1gi/6841176/Q96FW11.
8/(58.
54+/19.
8vs(32.
01+/14.
53)Pre-B2,t(12;21)p≤0.
050.
787Two-dimensionalgelelectrophoresis(2-DE)BatchesarelocatedFigure1A(incaseofthetopeleven)andAdditionalfile1:FigureS2(forthefollowing15th).
Incolumn'proteinnames,R1,R2andR3aretheMascotrankbasedonthewholeionicscoresdetected(upperorequalto48).
Poweranalysisisperformedindependentlyforeachspot,takingintoconsiderationthesamplesizeandvarianceexpressionvalue(ProgenesisSameSpotsV4).
CNN2=calponin-2;CDSα=cysteinedesulfurase,mitochondrialisoforma;PITPβ=phosphatidyl-inositoltransferproteinbetaisoform;BUB3a=mitoticcheckpointproteinBUB3isoforma;PDHE1=chainA,pyruvatedehydrogenaseE1S264eVariant;MAT2β=methionineadenosyltransferase2,subunitbeta,isoform1;PSMB2=proteasomesubunitbetatype-2,isoform1;CECR5=cateyesyndromecriticalregionprotein5isoform2;CK2α=caseinkinaseIIsubunitalphaisoforma;SEPT9_i3=MLLseptin-likefusionproteinMSF-B;hnRNPA2=heterogeneousnuclearribonucleo-proteinsA2;IVAD=ChainA,isovaleryl-Coadehydrogenase;FBA=fructosebisphosphatealdolase;PSMB6=proteasomesubunit6(p42);OTUB1=deubiquitinatingenzymeOTUB1.
Costaetal.
ClinicalProteomics2014,11:31Page4of12http://www.
clinicalproteomicsjournal.
com/content/11/1/31componentsanalysis(PCA)(seePatients,materialsandmethods)indicatedthattheamountofallthe26spotsofinterest,whengatheredtogether,coulddiscriminate2groupsofpatient,correlatingwiththepresenceorabsenceoft(12;21)(Figure4A,B).
Amongtheproteinsinvolvedincellcycleregula-tion,arethecalponin-2(CNN2)andmethionineade-nosyltransferase2beta(MAT2β)variant1,whicharehighlyexpressedinALLwitht(12;21),usuallyassociatedtogoodprognosis(p0.
01andp≤0.
05(n=15);B/Spotsdifferentiallyexpressedwithp≤0.
01(n=11).
NamesclosetodotesareidentificationbadgeofpatientslistedinTable1.
Costaetal.
ClinicalProteomics2014,11:31Page6of12http://www.
clinicalproteomicsjournal.
com/content/11/1/31Table3ConcludingremarksandfuturedirectionsExpressionobservedint(12;21)ALLsMajorexpectedcellulareffectsCommentsFuturedirectionsOverexpressionofCNN2MediatescellulargrowtharrestInfavorofgoodprognosisassociatedtot(12;21)IsCNN2amarkerofgoodprognosisIthastobeconfirmedwithalargestgroupofpatientsOverexpressionofMAT-2βFavorsthecellproliferationwithanti-apoptoticeffectWouldbeamarkerofcanceraggressivenessWouldbeexploreasatargetofchemotherapyOverexpressionofhnRNPA2FavorsthecellproliferationWouldbeamarkerofcanceraggressivenessWouldbeexploreasatargetofchemotherapyOverexpressionofPITPβSlowdownthecellproliferationIsitamarkerofgoodprognosisExplorationofthebalancePITPα/βUnderexpressionofBUB3AvoidslippageofmitosisOverexpressioninnont(12;21)couldleadtohypo/hyperpolyploidcellsMitosesanalysisUnderexpressionofhnRNPE2FavorsapoptosisInfavorofgoodprognosisassociatedtot(12;21)ExplorationofthebalancebetweenhnRNPE2,PSMB2andPSMB6and/orcheckuptheapoptosispathwaysOverexpressionofPSMB2FavorsapoptosisUnderexpressionofPSMB6HindersapoptosisInfavorofleukogenesisUnderexpressionofCK2αSlowdowntheproliferationbyapoptoticeffectInfavorofgoodprognosisassociatedtot(12;21)BothexpressionsincombinationwithIkaros(Ikzf1)phosphorylationanddegradationoughttobeclarifiedpreciselyOverexpressionofHSPC263(OTUB1)FavorsdeubiquitinationDoesitpreventIkaros(Ikzf1)degradationUnderexpressionofmetabolismpathwayproteins(i.
e.
,CDSα;CECR5;PDH;IVAD;IDH;Electrontransferflavoprotein)SignthelevelofenergeticconsumptionInfavorofgoodprognosisassociatedtot(12;21)Checkuplymphoblastsmetabolismpathway:HighamountwouldbeinaccordwithcanceraggressivenessCostaetal.
ClinicalProteomics2014,11:31Page7of12http://www.
clinicalproteomicsjournal.
com/content/11/1/31theregulatorysubunitoftheisozyme[15,17].
Whenex-pressionofMAT2βissuccessfullysilenced,excessiveapoptosisandgrowthslowdownisobservedinJurkatleukemicTcell[17,18].
RecentlyMAT2βv1(herechar-acterized),hasbeenidentifiedasananti-apoptoticcom-ponentthroughSirtuin1signaling[19].
Therefore,theoverexpressionofMAT2βint(12;21)ALLscannotbeconsideredasagoodprognosticfactor.
ItseemstohavetheoppositeeffectoncellproliferationwhencomparedtoCNN2.
MAT-2βhastobeevaluatedasapossibletargettoimproveprognosisinthet(12;21)ALLs(Table3).
HnRNPA2functionsastelomericcappingfactorprotect-ingtelomericDNAinvivoagainstnucleasedigestionbyrecruitingtheessentialRNAsubunit(hTR)tothetelomere[20].
RNAi,reductionofhnRNPA1andhnRNPA2signifi-cantlyreducedtheproliferationrateofColo16cells,sug-gestingthatthesehnRNPmembersmaybeclassifiedasoncogenes[21].
HnRNPA2isfoundoverexpressedint(12,21)ALLsandthuswouldcontributetofavorthepre-B2prolif-erationandwouldbeamarkerofleukemogenesis(Table3).
Thethreelastproteinsincludedinthisgroup(i.
e.
hnRNP-E1,BUB3αandSEPT9_i4)areunderexpressedint(12;21)ALLs.
HnRNPE1isamemberoftheRNAbindingproteinfam-ilywhichincludesimportantmitoticregulatorsandeffec-tors,havingmultiplefunctionsinhematopoieticcells.
Itisinvolvedincontrollingstemcellproliferationanddifferen-tiation[21,22].
Particularly,actingas"chaperone"moleculeontheRNA,hnRNPsE1functionsascrucialmodulatorofmRNAstability,andtranslationinhematopoieticcelldifferentiation[23].
Itstimulatesthetranslationofc-mycandBag-1,havingananti-apoptoticeffectoncancercells[21,24].
TheaccumulationofhnRNPE1inlymphoblastescouldinduceananti-apoptoticeffect,worseningthedisease.
TheunderexpressionofhnRNPE2observedint(12;21)ALLswouldcontributetoapoptosisintheseleukemiccells(Table3).
TheBUB3αisoneofthecomponentsofthespindleas-semblycheckpoint(SAC)whichinteractswithBUB1andleadstodelaythecellcycleprogression[25].
However,inconditionswheretheSACiskeptactive,somecellsescapefrommitosis,resultinginpolyploidcells[25].
Itsoverex-pressioninnont(12;21)ALLscouldcontributetotheslip-pageofmitosistowardahypo/polyploidcells(Table3).
SEPT9_i4isanisoformoftheGTPbindingseptinfamily,involvedincytokinesis.
OverexpressionoftheshortisoformSEPT9_i4disturbsseptininteractionsandcellularmotilitywhichcanberelatedtoneoplasicasso-ciatedphenotypes[26].
SEPT9_i4overexpressingcellshaveenhancedsurvivalinthepresenceofclinicallyrele-vantmicrotubuleactingdrugs.
Itsoverexpressioninseveralcancersmaybeclinicallyrelevantwithacontri-butionofdrugresistanceformsofcancer[27].
Thisi4variantistheCOOHterminalsequenceofalltheisoformes[28].
Therebyitmaybedifficulttoreachitasaspecifictarget.
ByintroducingmutationsthatpreventSEPT9self-associationattheN-andC-termini,ithasbeenshownthatseptinfilamentdisruptioncausesde-fectsinthelatestagesofcelldivisionbeforeabscission,similartotheseobserveduponSEPT9depletion[28].
ProteinsinvolvedinapoptosisAmongproteinswhichareprincipallyimplicatedinapop-toticpathwaysandmayhaveaprognosticvalue(i.
e.
,PITPβ,CK2α,andPSMB2,PSMB6),phosphatidylinositoltransferproteinsbeta(PITPβ)andCK2αhaveanimpactonPl3Kpathway.
PITPβwashighlyexpressedinthegroupoft(12;21)ALLs.
Physiologically,PITPβispoorlyexpressedandfa-vorstheactivationofagolgianPI3K[29].
PITPβaccu-mulation,bymodifyingthebalancePITPα/β,inducesadecreaseofproliferationandanincreaseofapoptosis[30].
TheexplorationofthebalancePITPα/βinALLsmaybeausefulbiologicalmarker.
Onthecontrary,CK2αwasunderexpressedint(12;21)ALLs.
CK2αdisturbsthePI3Kpathwaybyphos-phorylatingPTENandAKT,therebyfavoringcellprolif-erationbyananti-apoptoticeffect[7,31].
TheoverexpressionofCK2αhasbeenshowntoincreasethedeg-radationofIkarosprotein(i.
e.
,atumorsuppressorinALL)viatheubiquitinpathway[3,32,33].
Interestinglyinthet(12,21)ALLs,notonlyCK2αisunderexpressedbutOTUB1,adeubiquitinatingenzyme,isfoundslightlyoverexpressed(seeTable2,rank12)[34].
OTUB1hydrolasecanspecificallyremove'Lys-48"-linkedconju-gatedubiquitinfromproteinsandplaysanimportantregulatoryroleatthelevelofproteinturnoverbypre-ventingdegradation.
Ithasrecentlybeenidentifiedasanovelp53regulator[35].
AlltogetherthiscouldresultinanpreservationofIkarosfunctionsandwouldcontributetothegoodprognosticclaimedfort(12;21)ALLs.
Thetwootherproteinsimplicatedinapoptosis(i.
e.
,PSMB2andPSMB6)areconstitutiveoftheproteasome.
Theproteasomeisresponsibleforthedegradationofintracellularproteinsinvolvedincellcyclecontrolandregulationofapoptosis,includingthetumorsuppressorproteinp53andspecificcyclindependentkinases[36].
AtthistimewecannotexplainwhyPSMB2ispredominantint(12;21)ALLs,whilePSMB6isweaklyexpressed.
Newtherapiesagainsttumorgrowthanddevelopmenthaveananti-ubiquitin-proteasomepathwayeffect[36].
Twopathwaysinwhichmostoftheseproteins(i.
e.
,in-volvedinapoptosisandalsoBUB3)werepointedoutwithProteinCenterSoftware(ThermoScientificProteinCenterSoftware).
Thefirstone(Rawp-value,p0.
01andp≤0.
05)arenumbered.
NumberscorrespondtoproteinspotslistedinTable2.
FigureS3.
Mascotresearchresultsforthefirstelevenspotproteinsorderedbystatisticrank(ProgenesisSameSpotsV4software–seeAdditionalfile1:TableS1).
Foreachidentifiedtrypsicpeptide,individualionsscoresupto48indicateidentityorextensivehomology(p<0.
05).
Matchedpeptidesareshowninboldred.
Abbreviations2-DE:Two-dimensionalpolyacrylamidegelelectrophoresis;ALL:Acutelymphoblasticleukemia;CNN2:h2-calponin;MAT2Bβ:Methionineadenosyltransferase2β;hnRNP-E1:Heterogeneousnuclearribonucleoprotein-E1;BUB3α:MitoticcheckpointproteinBUB3isoforma;MSF-B:SEPT9_i4,MLLseptin-likefusionprotein;PITPβ:Phosphatidylinositoltransferproteinsbeta;CK2α:Caseinkinase2alpha;PSMB6:Proteasomesubunitsbeta2,PSMB2,proteasomesubunitsbeta6(p42);CDSα:Cysteindesulfurasemitochondrialisoformα;PDH:Pyruvatedehydrogenase;IVAD:IsovalerylCoAdehydrogenase.
CompetinginterestsTheauthorsdeclarethattheyhavenocompetinginterests.
OnlineSupplementaryinformationsareavailableatwww.
haematologica.
org.
Costaetal.
ClinicalProteomics2014,11:31Page10of12http://www.
clinicalproteomicsjournal.
com/content/11/1/31Authors'contributionsOC,PhD,MCU,performedanddesignedresearch,analyzeddata,andwrotethemanuscript;PS,MD,PUPH,ispediatrichematologistandwrotethemanuscript;EL,Technician,keepgrowingcelllines,DP,MD,performedcytogeneticanalysis,LC,PhD,RI,andPC,PhD,RI,performedmassspectrometryresearch,TJ,PhD,PU,DirectorofPISSAROProteomicfacility;MV,PUPHandJ-PV,MD,PUPHcontributedvitalreagents.
Allauthorsreadandapprovedthefinalmanuscript.
AcknowledgmentsTheauthorsthankFranoiseThouretforhertechnicalassistanceinleukemiacellspreparations.
Thisworkwassupportedbygrantsfrom:theLionsclubsfromNormandie(France),theFondationMartineMIDY(France)andtheAssociationLauretteFUGAIN,conventionALF/N°10-07(France).
Authordetails1LaboratoireMERCI,FacultédeMédecineetdePharmaciedeRouen,123boulevardGambetta,Rouen,Cedex76183,France.
2Serviced'Immuno-HématologieOnco-pédiatriqueduCHRUdeRouen,HpitalCharlesNicolle,Rouen76031,France.
3PISSAROProteomicfacility,(IRIB),U-Rouen,MontSaint-Aignan,France.
4CNRSUMR6270,TeamBiofilms,Résistance,InteractionsCellules-Surfaces,U-Rouen,MontSaint-Aignan,France.
5LaboratoiredeCytogénétique,CentreHenriBecquerel,Rouen76000,France.
Received:21January2014Accepted:14July2014Published:1August2014References1.
MullighanCG:Themoleculargeneticmakeupofacutelymphoblasticleukemia.
HematologyAmSocHematolEducProgram2012,2012:389–396.
Review.
2.
RomanaSP,PoirelH,LeconiatM,FlexorMA,MauchaufféM,JonveauxP,MacintyreEA,BergerR,BernardOA:Highfrequencyoft(12;21)inchildhoodB-lineageacutelymphoblasticleukemia.
Blood1995,86:4263–4269.
3.
LiZ,SongC,OuyangH,LaiL,PayneKJ,DovatS:Cellcycle-specificfunctionofIkarosinhumanleukemia.
PediatrBloodCancer2012,59:69–76.
4.
KornblauSM,TibesR,QiuYH,ChenW,KantarjianHM,AndreeffM,CoombesKR,MillsGB:FunctionalproteomicprofilingofAMLpredictsresponseandsurvival.
Blood2009,113:154–164.
5.
LuczakM,KamierczakM,HandschuhL,LewandowskiK,KomarnickiM,FiglerowiczM:Comparativeproteomeanalysisofacutemyeloidleukemiawithandwithoutmaturation.
JProteomics2012,75:5734–5748.
6.
MartinsLR,LúcioP,SilvaMC,GameiroP,SilvaMG,BarataJT:OnCK2regulationofchroniclymphocyticleukemiacellviability.
MolCellBiochem2011,356:51–55.
7.
BarataJT:TheimpactofPTENregulationbyCK2onPI3K-dependentsignalingandleukemiacellsurvival.
AdvEnzymeRegul2011,51:37–49.
8.
ScottM,HylandPL,McGregorG,HillanKJ,RussellSE,HallPA:MultimodalityexpressionprofilingshowsSEPT9tobeoverexpressedinawiderangeofhumantumours.
Oncogene2005,24:4688–4700.
9.
JiangN,KhamSK,KohGS,SuangLimJY,AriffinH,ChewFT,YeohAE:Identificationofprognosticproteinbiomarkersinchildhoodacutelymphoblasticleukemia(ALL).
JProteomics2011,74:843–857.
10.
GazyI,KupiecM:Theimportanceofbeingmodified:PCNAmodificationandDNAdamageresponse.
CellCycle2012,11:2620–2623.
11.
HossainMM,HwangDY,HuangQQ,SasakiY,JinJP:Developmentallyregulatedexpressionofcalponinisoformsandtheeffectofh2-calponinoncellproliferation.
AmJPhysiolCellPhysiol2003,284:C156–C167.
12.
WuKC,JinJP:Calponininnon-musclecells.
CellBiochemBiophys2008,52:139–148.
13.
YamamuraH,YoshikawaH,TatsutaM,AkedoH,TakahashiK:Expressionofthesmoothmusclecalponingeneinhumanosteosarcomaanditspossibleassociationwithprognosis.
IntJCancer1998,79:245–250.
14.
KatohY,IkuraT,HoshikawaY,TashiroS,ItoT,OhtaM,KeraY,NodaT,IgarashiK:MethionineadenosyltransferaseIIservesasatranscriptionalcorepressorofMafoncoprotein.
MolCell2011,41:554–566.
15.
NordgrenKK,PengY,PelleymounterLL,MoonI,AboR,FengQ,EckloffB,YeeVC,WiebenE,WeinshilboumRM:Methionineadenosyltransferase2A/2Bandmethylation:genesequencevariationandfunctionalgenomics.
DrugMetabDispos2011,39:2135–2147.
16.
WangQ,LiuQY,LiuZS,QianQ,SunQ,PanDY:InhibitionofhepatocelluarcarcinomaMAT2AandMAT2betageneexpressionsbysingleanddualsmallinterferingRNA.
JExpClinCancerRes2008,27:72–80.
17.
JaniTS,GobejishviliL,HotePT,BarveAS,Joshi-BarveS,KharebavaG,SuttlesJ,ChenT,McClainCJ,BarveS:InhibitionofmethionineadenosyltransferaseIIinducesFasLexpression,Fas-DISCformationandcaspase-8-dependentapoptoticdeathinTleukemiccells.
CellRes2009,19:358–369.
18.
AttiaRR,GardnerLA,MahrousE,TaxmanDJ,LegrosL,RoweS,TingJP,GellerA,KotbM:SelectivetargetingofleukemiccellgrowthinvivoandinvitrousingagenesilencingapproachtodiminishS-adenosylmethioninesynthesis.
JBiolChem2008,283:30788–30795.
19.
YangH,ZhengY,LiTW,PengH,FernandezRamosD,Martínez-ChantarML,RojasAL,MatoJM,LuSC:Methionineadenosyltransferase2B,HuRandsirtuin1crosstalkimpactsonResveratrol'seffectonapoptosisandgrowthinlivercancercells.
JBiolChem2013,288:23161–23170.
20.
Moran-JonesK,WaymanL,KennedyDD,ReddelRR,SaraS,SneeMJ,SmithR:HnRNPA2apotentialssDNA/RNAmolecularadapteratthetelomere.
NucleicAcidsRes2005,33:486–496.
21.
CarpenterB,MacKayC,AlnabulsiA,MacKayM,TelferC,MelvinWT,MurrayGI:Therolesofheterogeneousnuclearribonucleoproteinsintumourdevelopmentandprogression.
BiochimBiophysActa2006,1765:85–100.
22.
ChaudhuryA,ChanderP,HowePH:Heterogeneousnuclearribonucleoproteins(hnRNPs)incellularprocesses:FocusonhnRNPE1'smultifunctionalregulatoryroles.
RNA2010,16:1449–1462.
Review.
23.
Ostareck-LedererA,OstareckDH:Precisionmechanicswithmultifunctionaltools:howhnRNPKandhnRNPsE1/E2contributetopost-transcriptionalcontrolofgeneexpressioninhematopoiesis.
CurrProteinPeptSci2012,13:391–400.
24.
WangH,YeY,PanSY,ZhuGY,LiYW,FongDW,YuZL:ProteomicidentificationofproteinsinvolvedintheanticanceractivitiesoforidonininHepG2cells.
Phytomedicine2011,18:163–169.
25.
NiikuraY,OgiH,KikuchiK,KitagawaK:BUB3thatdissociatesfromBUB1activatescaspase-independentmitoticdeath(CIMD).
CellDeathDiffer2010,17:1011–1024.
26.
RobertsonC,ChurchSW,NagarHA,PriceJ,HallPA,RussellSE:PropertiesofSEPT9isoformsandtherequirementforGTPbinding.
JPathol2004,203:519–527.
27.
ChackoAD,McDadeSS,ChanduloyS,ChurchSW,KennedyR,PriceJ,HallPA,RussellSE:ExpressionoftheSEPT9_i4isoformconfersresistancetomicrotubule-interactingdrugs.
CellOncol2012,35:85–93.
28.
KimMS,FroeseCD,EsteyMP,TrimbleWS:SEPT9occupiestheterminalpositionsinseptinoctamersandmediatespolymerization-dependentfunctionsinabscission.
JCellBiol2011,195:815–826.
29.
CarvouN,HolicR,LiM,FutterC,SkippenA,CockcroftS:Phosphatidylinositol-andphosphatidylcholine-transferactivityofPITP-betaisessentialforCOPI-mediatedretrogradetransportfromtheGolgitotheendoplasmicreticulum.
JCellSci2010,123:1262–1273.
30.
SnoekGT:Phosphatidylinositoltransferproteins:emergingrolesincellproliferation,celldeathandsurvival.
IUBMBLife2004,56:467–475.
Review.
31.
RimanS,RizkallahR,KassardjianA,AlexanderKE,LüscherB,HurtMM:PhosphorylationofthetranscriptionfactorYY1byCK2αpreventscleavagebycaspase7duringapoptosis.
MolCellBiol2012,32:797–807.
32.
PayneKJ,DovatS:Ikarosandtumorsuppressioninacutelymphoblasticleukemia.
CritRevOncog2011,16:3–12.
Review.
33.
DovatS,SongC,PayneKJ,LiZ:Ikaros,CK2kinase,andtheroadtoleukemia.
MolCellBiochem2011,356:201–207.
Review.
34.
WienerR,ZhangX,WangT,WolbergerC:ThemechanismofOTUB1-mediatedinhibitionofubiquitination.
Nature2012,483:618–622.
35.
SunXX,ChallagundlaKB,DaiMS:Positiveregulationofp53stabilityandactivitybythedeubiquitinatingenzymeOtubain1.
EMBOJ2012,31:576–592.
36.
LudwigH,KhayatD,GiacconeG,FaconT:Proteasomeinhibitionanditsclinicalprospectsinthetreatmentofhematologicandsolidmalignancies.
Cancer2005,104:1794–1797.
37.
DavisCD,TsujiPA,MilnerJA:Selenoproteinsandcancerprevention.
AnnuRevNutr2012,32:73–95.
Review.
38.
CarletM,JanjetovicK,RainerJ,SchmidtS,Panzer-GrümayerR,MannG,PrelogM,MeisterB,PlonerC,KoflerR:Expression,regulationandfunctionofphosphofructo-kinase/fructose-biphosphatases(PFKFBs)inglucocorticoid-inducedapoptosisofacutelymphoblasticleukemiacells.
BMCCancer2010,10:638–649.
Costaetal.
ClinicalProteomics2014,11:31Page11of12http://www.
clinicalproteomicsjournal.
com/content/11/1/3139.
KowalskiW,NoconD,GamianA,KoodziejJ,RakusD:AssociationofC-terminalregionofphosphoglyceratemutasewithglycolyticcomplexregulatesenergyproductionincancercells.
JCellPhysiol2012,227:2613–2621.
40.
HullemanE,KazemierKM,HollemanA,VanderWeeleDJ,RudinCM,BroekhuisMJ,EvansWE,PietersR,DenBoerML:Inhibitionofglycolysismodulatesprednisoloneresistanceinacutelymphoblasticleukemiacells.
Blood2009,113:2014–2021.
41.
FootzTK,Brinkman-MillsP,BantingGS,MaierSA,RiaziMA,BridglandL,HuS,BirrenB,MinoshimaS,ShimizuN,PanH,NguyenT,FangF,FuY,RayL,WuH,ShaullS,PhanS,YaoZ,ChenF,HuanA,HuP,WangQ,LohP,QiS,RoeBA,McDermidHE:Analysisofthecateyesyndromecriticalregioninhumansandtheregionofconservedsyntenyinmice:asearchforcandidategenesatornearthehumanchromosome22pericentromere.
GenomeRes2001,11:1053–1070.
42.
MichaudJ,SimpsonKM,EscherR,Buchet-PoyauK,BeissbarthT,CarmichaelC,RitchieME,SchützF,CannonP,LiuM,ShenX,ItoY,RaskindWH,HorwitzMS,OsatoM,TurnerDR,SpeedTP,KavallarisM,SmythGK,ScottHS:IntegrativeanalysisofRUNX1downstreampathwaysandtargetgenes.
BMCGenomics2008,9:363–380.
43.
JiangN,KohGS,LimJY,KhamSK,AriffinH,ChewFT,YeohAE:BIMisaprognosticbiomarkerforearlyprednisoloneresponseinpediatricacutelymphoblasticleukemia.
ExpHematol2011,39:321–329.
44.
TrembleyJH,ChenZ,UngerG,SlatonJ,KrenBT,VanWaesC,AhmedK:EmergenceofproteinkinaseCK2asakeytargetincancertherapy.
Biofactors2010,36:187–195.
45.
PolakR,BuitenhuisM:ThePI3K/PKBsignalingmoduleaskeyregulatorofhematopoiesis:implicationsfortherapeuticstrategiesinleukemia.
Blood2012,119:911–923.
46.
ErnstA,AvvakumovG,TongJ,FanY,ZhaoY,AlbertsP,PersaudA,WalkerJR,NeculaiAM,NeculaiD,VorobyovA,GargP,BeattyL,ChanPK,JuangYC,LandryMC,YehC,ZeqirajE,KaramboulasK,Allali-HassaniA,VedadiM,TyersM,MoffatJ,SicheriF,PelletierL,DurocherD,RaughtB,RotinD,YangJ,MoranMF,etal:Astrategyformodulationofenzymesintheubiquitinsystem.
Science2013,339:590–595.
47.
SchneiderP,CostaO,LegrandE,BigotD,LecleireS,GrassiV,VannierJP,VasseM:Invitrosecretionofmatrixmetalloprotease9isaprognosticmarkerinchildhoodacutelymphoblasticleukemia.
LeukRes2010,34:24–31.
48.
GandemerV,ChevretS,PetitA,VermylenC,LeblancT,MichelG,SchmittC,LejarsO,SchneiderP,DemeocqF,Bader-MeunierB,BernaudinF,PerelY,AuclercMF,CayuelaJM,LevergerG,BaruchelA,FRALLEGroup:ExcellentprognosisoflaterelapsesofETV6/RUNX1-positivechildhoodacutelymphoblasticleukemia:lessonsfromtheFRALLE93protocol.
Haematologica2012,97:1743–1750.
49.
VanDongenJJ,MacintyreEA,GabertJA,DelabesseE,RossiV,SaglioG,GottardiE,RambaldiA,DottiG,GriesingerF,ParreiraA,GameiroP,DiázMG,MalecM,LangerakAW,SanMiguelJF,BiondiA:StandardizedRT-PCRanalysisoffusiongenetranscriptsfromchromosomeaberrationsinacuteleukemiafordetectionofminimalresidualdisease.
ReportoftheBIOMED-1ConcertedAction:investigationofminimalresidualdiseaseinacuteleukemia.
Leukemia1999,13:1901–1928.
50.
SchneiderP,VasseM,CorbièreC,LegrandE,Marie-CardineA,BoquetC,VannierJP:Endostatinvariationsinchildhoodacutelymphoblasticleukaemia-comparisonwithbasicfibroblastgrowthfactorandvascularendothelialgrowthfactor.
LeukRes2007,31:629–638.
51.
TnaniH,LópezI,JouenneT,VicientaCM:Proteincompositionanalysisofoilbodiesfrommaizeembryosduringgermination.
JPlantPhysiol2011,168:510–513.
52.
MagdeldinS,EnanyS,YoshidaY,XuB,ZhangY,ZureenaZ,LokamaniI,YaoitaE,YamamotoT:Basicsandrecentadvancesoftwodimensional-polyacrylamidegelelectrophoresis.
ClinProteomics2014,11:16–26.
doi:10.
1186/1559-0275-11-31Citethisarticleas:Costaetal.
:Proteomicprofileofpre-B2lymphoblastsfromchildrenwithacutelymphoblasticleukemia(ALL)inrelationwiththetranslocation(12;21).
ClinicalProteomics201411:31.
SubmityournextmanuscripttoBioMedCentralandtakefulladvantageof:ConvenientonlinesubmissionThoroughpeerreviewNospaceconstraintsorcolorgurechargesImmediatepublicationonacceptanceInclusioninPubMed,CAS,ScopusandGoogleScholarResearchwhichisfreelyavailableforredistributionSubmityourmanuscriptatwww.
biomedcentral.
com/submitCostaetal.
ClinicalProteomics2014,11:31Page12of12http://www.
clinicalproteomicsjournal.
com/content/11/1/31
InthelightofproteinmarkerstudiestoclassifysubtypesofAcuteMyeloblasticLeukemiaefficiently,wehavecomparedthelymphoblastesproteomeinChildhoodALLinaccordancewiththepresenceoft(12;21),indicatorofgoodprognosis,usually.
Methods:Proteinexpressioninpre-B2lymphoblasticcells,collectedfromresidualbonemarrowcellsafterdiagnosticprocedures,wasanalyzedusingtwodimensionalgelelectrophoresisprotocol.
Proteinspotswhoseaveragenormalizedvolumeswerestatisticallydifferentinthetwopatientsgroups(n=13;studentttestp48)(seeOnlineAdditionalfile1:FigureS3)wereofgreatinterestintermsofleukemogenesissincein-volvedinmechanismsrelatedtoeitherthecellcycleregu-lation,orapoptosis,orenergeticmetabolism(i.
e.
,43%,29%or28%ofthem,respectively)(Figure2B).
PrincipalCostaetal.
ClinicalProteomics2014,11:31Page2of12http://www.
clinicalproteomicsjournal.
com/content/11/1/31Table1Clinicalandbiologicalstatusofchildhoodpre-B2ALLpatientsGroupofALLpatientsPatientsIDAgeatdiagnosis(Months)Peripheralbloodlymphocytes(Giga/L)KaryotypeFusiontranscriptsOutcomeWitht(12;21)BACYO2448,7t(12;21)ETV6/RUNX1CR1GUEKE11842,2t(12;21)ETV6/RUNX1CR1LECRO765,4t(12;21)ETV6/RUNX1CR1MARLE42166t(12;21)ETV6/RUNX1CR1PELAD5020t(12;21)ETV6/RUNX1Dead*RIFEM76216t(12;21)ETV6/RUNX1CR1OtherkaryotypeabnormalitiesBOILE182,8dic(9;20)NACR1CREBA18011,6mono21NACR1DESNA1315,5rear9pNACR1JUGEL348,5HyperploidyNACR1LEMAL4856t(9;22)BCL/ABL1CR1MORLU171171t(9;22)BCL/ABL1Dead*ROSLO576,1HyperploidyNACR1TheboldnumbersindicatesvalueshigherthethresholdofFRALLE2000protocol.
CR1=Firstcompleteremission;dic=dicentric;ID=Identification;mono=monosomy;NA=Notapplicable;rear=Rearrangement.
*=DirectlyrelatedtotherecurrenceofALL.
Figure1Differentialexpressionofproteinsinbonemarrowlymphoblasticcellsfromchildhoodpre-BALL.
–A–Down:Two-DEgelimageoflymphoblasticcellsproteinsissuingfrombonemarrow.
NumbersrefertothetopelevenproteinswhichhavebeencharacterizedandlistedinTable1b.
Up:Magnificationviewofinterestingzone–B–3Dmontageofthethreetopdifferentiallyexpressedspotproteins(i.
e.
,2-DEBatch2768,3185and4083onrank1,2and3,respectively).
Only2individualspotimagesineachgroup(1and2=Pre-B2ALLwitht(12;21),1′and2′=Pre-B2ALLwithotherkaryotypeabnormalities)areshown.
Costaetal.
ClinicalProteomics2014,11:31Page3of12http://www.
clinicalproteomicsjournal.
com/content/11/1/31Table2Theclassificationofdifferentiallyexpressedproteinsinpre-B2lymphoblasticcellsfromchildrenwithALLSpots:statisticrank(2-DEbatch)Proteinnames[Homosapiens]NCBIanduniprotIDnumbersFoldchanges/ALLt(12;21)versusothers(averagenormalizedvolume+/SD)105GroupwiththeupperaveragevolumeStudent-ttestPower(afterqvalue)1(2768)CNN2gi/49456619/Q994391.
9/(38.
59+/8.
1vs20.
76+/4)Pre-B2,t(12;21)p≤0.
0010.
9982(3185)CDSαgi/32307132/Q9Y697-12.
2/(9.
5+/3.
8vs20.
98+/5.
2)OtherPre-B2p≤0.
0010.
9853(4083)PITPβgi/6912594/P487392/(12.
8+/3.
1vs6.
28+/2.
21)Pre-B2,t(12;21)p≤0.
0010.
9834(2457)hnRNP-E1gi/460771/Q153651.
4/(29.
87+/4.
69vs41.
57+/4.
88)OtherPre-B2p≤0.
0050.
976BUB3αgi/4757880/O436845(4069)PDH-E1gi/149242791/P13804-11.
5/(15.
16+/4.
33vs23.
29+/3.
12)OtherPre-B2p≤0.
0050.
9556(2090)MAT2βi1gi/11034825/Q9NZL91.
5/(21.
94+/3.
89vs15.
06+/2.
91)Pre-B2,t(12;21)p≤0.
0050.
9157(2800)PSMB2gi/4506195/B7Z4781.
7/(9.
48+/2.
49vs5.
69+/1.
63)Pre-B2,t(12;21)p≤0.
010.
8788(4081)CECR5gi/14861834/Q9BXW71.
3/(28.
51+/4.
48vs37.
3+/4.
81)OtherPre-B2p≤0.
010.
867BUB3αgi/4757880/O436849(3121)CK2αgi/4503095/P684001.
5/(13.
55+/2.
36vs20.
31+/4.
9)OtherPre-B2p≤0.
010.
864SEPT9i3gi/668381/Q9UHD8-310(2056)HnRNPA2gi/4504447/P22626-22/(18.
28+/6.
56vs9.
26+/4.
77)Pre-B2,t(12;21)p≤0.
010.
85411(3120)IVADgi/3212539/P264401.
5/(14.
98+/3.
44vs22.
64+/5.
34)OtherPreB2p≤0.
010.
835FBAgi/312137/P05062PSMB6gi/1526426/P6233312(2656)OTUB1gi/6841176/Q96FW11.
8/(58.
54+/19.
8vs(32.
01+/14.
53)Pre-B2,t(12;21)p≤0.
050.
787Two-dimensionalgelelectrophoresis(2-DE)BatchesarelocatedFigure1A(incaseofthetopeleven)andAdditionalfile1:FigureS2(forthefollowing15th).
Incolumn'proteinnames,R1,R2andR3aretheMascotrankbasedonthewholeionicscoresdetected(upperorequalto48).
Poweranalysisisperformedindependentlyforeachspot,takingintoconsiderationthesamplesizeandvarianceexpressionvalue(ProgenesisSameSpotsV4).
CNN2=calponin-2;CDSα=cysteinedesulfurase,mitochondrialisoforma;PITPβ=phosphatidyl-inositoltransferproteinbetaisoform;BUB3a=mitoticcheckpointproteinBUB3isoforma;PDHE1=chainA,pyruvatedehydrogenaseE1S264eVariant;MAT2β=methionineadenosyltransferase2,subunitbeta,isoform1;PSMB2=proteasomesubunitbetatype-2,isoform1;CECR5=cateyesyndromecriticalregionprotein5isoform2;CK2α=caseinkinaseIIsubunitalphaisoforma;SEPT9_i3=MLLseptin-likefusionproteinMSF-B;hnRNPA2=heterogeneousnuclearribonucleo-proteinsA2;IVAD=ChainA,isovaleryl-Coadehydrogenase;FBA=fructosebisphosphatealdolase;PSMB6=proteasomesubunit6(p42);OTUB1=deubiquitinatingenzymeOTUB1.
Costaetal.
ClinicalProteomics2014,11:31Page4of12http://www.
clinicalproteomicsjournal.
com/content/11/1/31componentsanalysis(PCA)(seePatients,materialsandmethods)indicatedthattheamountofallthe26spotsofinterest,whengatheredtogether,coulddiscriminate2groupsofpatient,correlatingwiththepresenceorabsenceoft(12;21)(Figure4A,B).
Amongtheproteinsinvolvedincellcycleregula-tion,arethecalponin-2(CNN2)andmethionineade-nosyltransferase2beta(MAT2β)variant1,whicharehighlyexpressedinALLwitht(12;21),usuallyassociatedtogoodprognosis(p0.
01andp≤0.
05(n=15);B/Spotsdifferentiallyexpressedwithp≤0.
01(n=11).
NamesclosetodotesareidentificationbadgeofpatientslistedinTable1.
Costaetal.
ClinicalProteomics2014,11:31Page6of12http://www.
clinicalproteomicsjournal.
com/content/11/1/31Table3ConcludingremarksandfuturedirectionsExpressionobservedint(12;21)ALLsMajorexpectedcellulareffectsCommentsFuturedirectionsOverexpressionofCNN2MediatescellulargrowtharrestInfavorofgoodprognosisassociatedtot(12;21)IsCNN2amarkerofgoodprognosisIthastobeconfirmedwithalargestgroupofpatientsOverexpressionofMAT-2βFavorsthecellproliferationwithanti-apoptoticeffectWouldbeamarkerofcanceraggressivenessWouldbeexploreasatargetofchemotherapyOverexpressionofhnRNPA2FavorsthecellproliferationWouldbeamarkerofcanceraggressivenessWouldbeexploreasatargetofchemotherapyOverexpressionofPITPβSlowdownthecellproliferationIsitamarkerofgoodprognosisExplorationofthebalancePITPα/βUnderexpressionofBUB3AvoidslippageofmitosisOverexpressioninnont(12;21)couldleadtohypo/hyperpolyploidcellsMitosesanalysisUnderexpressionofhnRNPE2FavorsapoptosisInfavorofgoodprognosisassociatedtot(12;21)ExplorationofthebalancebetweenhnRNPE2,PSMB2andPSMB6and/orcheckuptheapoptosispathwaysOverexpressionofPSMB2FavorsapoptosisUnderexpressionofPSMB6HindersapoptosisInfavorofleukogenesisUnderexpressionofCK2αSlowdowntheproliferationbyapoptoticeffectInfavorofgoodprognosisassociatedtot(12;21)BothexpressionsincombinationwithIkaros(Ikzf1)phosphorylationanddegradationoughttobeclarifiedpreciselyOverexpressionofHSPC263(OTUB1)FavorsdeubiquitinationDoesitpreventIkaros(Ikzf1)degradationUnderexpressionofmetabolismpathwayproteins(i.
e.
,CDSα;CECR5;PDH;IVAD;IDH;Electrontransferflavoprotein)SignthelevelofenergeticconsumptionInfavorofgoodprognosisassociatedtot(12;21)Checkuplymphoblastsmetabolismpathway:HighamountwouldbeinaccordwithcanceraggressivenessCostaetal.
ClinicalProteomics2014,11:31Page7of12http://www.
clinicalproteomicsjournal.
com/content/11/1/31theregulatorysubunitoftheisozyme[15,17].
Whenex-pressionofMAT2βissuccessfullysilenced,excessiveapoptosisandgrowthslowdownisobservedinJurkatleukemicTcell[17,18].
RecentlyMAT2βv1(herechar-acterized),hasbeenidentifiedasananti-apoptoticcom-ponentthroughSirtuin1signaling[19].
Therefore,theoverexpressionofMAT2βint(12;21)ALLscannotbeconsideredasagoodprognosticfactor.
ItseemstohavetheoppositeeffectoncellproliferationwhencomparedtoCNN2.
MAT-2βhastobeevaluatedasapossibletargettoimproveprognosisinthet(12;21)ALLs(Table3).
HnRNPA2functionsastelomericcappingfactorprotect-ingtelomericDNAinvivoagainstnucleasedigestionbyrecruitingtheessentialRNAsubunit(hTR)tothetelomere[20].
RNAi,reductionofhnRNPA1andhnRNPA2signifi-cantlyreducedtheproliferationrateofColo16cells,sug-gestingthatthesehnRNPmembersmaybeclassifiedasoncogenes[21].
HnRNPA2isfoundoverexpressedint(12,21)ALLsandthuswouldcontributetofavorthepre-B2prolif-erationandwouldbeamarkerofleukemogenesis(Table3).
Thethreelastproteinsincludedinthisgroup(i.
e.
hnRNP-E1,BUB3αandSEPT9_i4)areunderexpressedint(12;21)ALLs.
HnRNPE1isamemberoftheRNAbindingproteinfam-ilywhichincludesimportantmitoticregulatorsandeffec-tors,havingmultiplefunctionsinhematopoieticcells.
Itisinvolvedincontrollingstemcellproliferationanddifferen-tiation[21,22].
Particularly,actingas"chaperone"moleculeontheRNA,hnRNPsE1functionsascrucialmodulatorofmRNAstability,andtranslationinhematopoieticcelldifferentiation[23].
Itstimulatesthetranslationofc-mycandBag-1,havingananti-apoptoticeffectoncancercells[21,24].
TheaccumulationofhnRNPE1inlymphoblastescouldinduceananti-apoptoticeffect,worseningthedisease.
TheunderexpressionofhnRNPE2observedint(12;21)ALLswouldcontributetoapoptosisintheseleukemiccells(Table3).
TheBUB3αisoneofthecomponentsofthespindleas-semblycheckpoint(SAC)whichinteractswithBUB1andleadstodelaythecellcycleprogression[25].
However,inconditionswheretheSACiskeptactive,somecellsescapefrommitosis,resultinginpolyploidcells[25].
Itsoverex-pressioninnont(12;21)ALLscouldcontributetotheslip-pageofmitosistowardahypo/polyploidcells(Table3).
SEPT9_i4isanisoformoftheGTPbindingseptinfamily,involvedincytokinesis.
OverexpressionoftheshortisoformSEPT9_i4disturbsseptininteractionsandcellularmotilitywhichcanberelatedtoneoplasicasso-ciatedphenotypes[26].
SEPT9_i4overexpressingcellshaveenhancedsurvivalinthepresenceofclinicallyrele-vantmicrotubuleactingdrugs.
Itsoverexpressioninseveralcancersmaybeclinicallyrelevantwithacontri-butionofdrugresistanceformsofcancer[27].
Thisi4variantistheCOOHterminalsequenceofalltheisoformes[28].
Therebyitmaybedifficulttoreachitasaspecifictarget.
ByintroducingmutationsthatpreventSEPT9self-associationattheN-andC-termini,ithasbeenshownthatseptinfilamentdisruptioncausesde-fectsinthelatestagesofcelldivisionbeforeabscission,similartotheseobserveduponSEPT9depletion[28].
ProteinsinvolvedinapoptosisAmongproteinswhichareprincipallyimplicatedinapop-toticpathwaysandmayhaveaprognosticvalue(i.
e.
,PITPβ,CK2α,andPSMB2,PSMB6),phosphatidylinositoltransferproteinsbeta(PITPβ)andCK2αhaveanimpactonPl3Kpathway.
PITPβwashighlyexpressedinthegroupoft(12;21)ALLs.
Physiologically,PITPβispoorlyexpressedandfa-vorstheactivationofagolgianPI3K[29].
PITPβaccu-mulation,bymodifyingthebalancePITPα/β,inducesadecreaseofproliferationandanincreaseofapoptosis[30].
TheexplorationofthebalancePITPα/βinALLsmaybeausefulbiologicalmarker.
Onthecontrary,CK2αwasunderexpressedint(12;21)ALLs.
CK2αdisturbsthePI3Kpathwaybyphos-phorylatingPTENandAKT,therebyfavoringcellprolif-erationbyananti-apoptoticeffect[7,31].
TheoverexpressionofCK2αhasbeenshowntoincreasethedeg-radationofIkarosprotein(i.
e.
,atumorsuppressorinALL)viatheubiquitinpathway[3,32,33].
Interestinglyinthet(12,21)ALLs,notonlyCK2αisunderexpressedbutOTUB1,adeubiquitinatingenzyme,isfoundslightlyoverexpressed(seeTable2,rank12)[34].
OTUB1hydrolasecanspecificallyremove'Lys-48"-linkedconju-gatedubiquitinfromproteinsandplaysanimportantregulatoryroleatthelevelofproteinturnoverbypre-ventingdegradation.
Ithasrecentlybeenidentifiedasanovelp53regulator[35].
AlltogetherthiscouldresultinanpreservationofIkarosfunctionsandwouldcontributetothegoodprognosticclaimedfort(12;21)ALLs.
Thetwootherproteinsimplicatedinapoptosis(i.
e.
,PSMB2andPSMB6)areconstitutiveoftheproteasome.
Theproteasomeisresponsibleforthedegradationofintracellularproteinsinvolvedincellcyclecontrolandregulationofapoptosis,includingthetumorsuppressorproteinp53andspecificcyclindependentkinases[36].
AtthistimewecannotexplainwhyPSMB2ispredominantint(12;21)ALLs,whilePSMB6isweaklyexpressed.
Newtherapiesagainsttumorgrowthanddevelopmenthaveananti-ubiquitin-proteasomepathwayeffect[36].
Twopathwaysinwhichmostoftheseproteins(i.
e.
,in-volvedinapoptosisandalsoBUB3)werepointedoutwithProteinCenterSoftware(ThermoScientificProteinCenterSoftware).
Thefirstone(Rawp-value,p0.
01andp≤0.
05)arenumbered.
NumberscorrespondtoproteinspotslistedinTable2.
FigureS3.
Mascotresearchresultsforthefirstelevenspotproteinsorderedbystatisticrank(ProgenesisSameSpotsV4software–seeAdditionalfile1:TableS1).
Foreachidentifiedtrypsicpeptide,individualionsscoresupto48indicateidentityorextensivehomology(p<0.
05).
Matchedpeptidesareshowninboldred.
Abbreviations2-DE:Two-dimensionalpolyacrylamidegelelectrophoresis;ALL:Acutelymphoblasticleukemia;CNN2:h2-calponin;MAT2Bβ:Methionineadenosyltransferase2β;hnRNP-E1:Heterogeneousnuclearribonucleoprotein-E1;BUB3α:MitoticcheckpointproteinBUB3isoforma;MSF-B:SEPT9_i4,MLLseptin-likefusionprotein;PITPβ:Phosphatidylinositoltransferproteinsbeta;CK2α:Caseinkinase2alpha;PSMB6:Proteasomesubunitsbeta2,PSMB2,proteasomesubunitsbeta6(p42);CDSα:Cysteindesulfurasemitochondrialisoformα;PDH:Pyruvatedehydrogenase;IVAD:IsovalerylCoAdehydrogenase.
CompetinginterestsTheauthorsdeclarethattheyhavenocompetinginterests.
OnlineSupplementaryinformationsareavailableatwww.
haematologica.
org.
Costaetal.
ClinicalProteomics2014,11:31Page10of12http://www.
clinicalproteomicsjournal.
com/content/11/1/31Authors'contributionsOC,PhD,MCU,performedanddesignedresearch,analyzeddata,andwrotethemanuscript;PS,MD,PUPH,ispediatrichematologistandwrotethemanuscript;EL,Technician,keepgrowingcelllines,DP,MD,performedcytogeneticanalysis,LC,PhD,RI,andPC,PhD,RI,performedmassspectrometryresearch,TJ,PhD,PU,DirectorofPISSAROProteomicfacility;MV,PUPHandJ-PV,MD,PUPHcontributedvitalreagents.
Allauthorsreadandapprovedthefinalmanuscript.
AcknowledgmentsTheauthorsthankFranoiseThouretforhertechnicalassistanceinleukemiacellspreparations.
Thisworkwassupportedbygrantsfrom:theLionsclubsfromNormandie(France),theFondationMartineMIDY(France)andtheAssociationLauretteFUGAIN,conventionALF/N°10-07(France).
Authordetails1LaboratoireMERCI,FacultédeMédecineetdePharmaciedeRouen,123boulevardGambetta,Rouen,Cedex76183,France.
2Serviced'Immuno-HématologieOnco-pédiatriqueduCHRUdeRouen,HpitalCharlesNicolle,Rouen76031,France.
3PISSAROProteomicfacility,(IRIB),U-Rouen,MontSaint-Aignan,France.
4CNRSUMR6270,TeamBiofilms,Résistance,InteractionsCellules-Surfaces,U-Rouen,MontSaint-Aignan,France.
5LaboratoiredeCytogénétique,CentreHenriBecquerel,Rouen76000,France.
Received:21January2014Accepted:14July2014Published:1August2014References1.
MullighanCG:Themoleculargeneticmakeupofacutelymphoblasticleukemia.
HematologyAmSocHematolEducProgram2012,2012:389–396.
Review.
2.
RomanaSP,PoirelH,LeconiatM,FlexorMA,MauchaufféM,JonveauxP,MacintyreEA,BergerR,BernardOA:Highfrequencyoft(12;21)inchildhoodB-lineageacutelymphoblasticleukemia.
Blood1995,86:4263–4269.
3.
LiZ,SongC,OuyangH,LaiL,PayneKJ,DovatS:Cellcycle-specificfunctionofIkarosinhumanleukemia.
PediatrBloodCancer2012,59:69–76.
4.
KornblauSM,TibesR,QiuYH,ChenW,KantarjianHM,AndreeffM,CoombesKR,MillsGB:FunctionalproteomicprofilingofAMLpredictsresponseandsurvival.
Blood2009,113:154–164.
5.
LuczakM,KamierczakM,HandschuhL,LewandowskiK,KomarnickiM,FiglerowiczM:Comparativeproteomeanalysisofacutemyeloidleukemiawithandwithoutmaturation.
JProteomics2012,75:5734–5748.
6.
MartinsLR,LúcioP,SilvaMC,GameiroP,SilvaMG,BarataJT:OnCK2regulationofchroniclymphocyticleukemiacellviability.
MolCellBiochem2011,356:51–55.
7.
BarataJT:TheimpactofPTENregulationbyCK2onPI3K-dependentsignalingandleukemiacellsurvival.
AdvEnzymeRegul2011,51:37–49.
8.
ScottM,HylandPL,McGregorG,HillanKJ,RussellSE,HallPA:MultimodalityexpressionprofilingshowsSEPT9tobeoverexpressedinawiderangeofhumantumours.
Oncogene2005,24:4688–4700.
9.
JiangN,KhamSK,KohGS,SuangLimJY,AriffinH,ChewFT,YeohAE:Identificationofprognosticproteinbiomarkersinchildhoodacutelymphoblasticleukemia(ALL).
JProteomics2011,74:843–857.
10.
GazyI,KupiecM:Theimportanceofbeingmodified:PCNAmodificationandDNAdamageresponse.
CellCycle2012,11:2620–2623.
11.
HossainMM,HwangDY,HuangQQ,SasakiY,JinJP:Developmentallyregulatedexpressionofcalponinisoformsandtheeffectofh2-calponinoncellproliferation.
AmJPhysiolCellPhysiol2003,284:C156–C167.
12.
WuKC,JinJP:Calponininnon-musclecells.
CellBiochemBiophys2008,52:139–148.
13.
YamamuraH,YoshikawaH,TatsutaM,AkedoH,TakahashiK:Expressionofthesmoothmusclecalponingeneinhumanosteosarcomaanditspossibleassociationwithprognosis.
IntJCancer1998,79:245–250.
14.
KatohY,IkuraT,HoshikawaY,TashiroS,ItoT,OhtaM,KeraY,NodaT,IgarashiK:MethionineadenosyltransferaseIIservesasatranscriptionalcorepressorofMafoncoprotein.
MolCell2011,41:554–566.
15.
NordgrenKK,PengY,PelleymounterLL,MoonI,AboR,FengQ,EckloffB,YeeVC,WiebenE,WeinshilboumRM:Methionineadenosyltransferase2A/2Bandmethylation:genesequencevariationandfunctionalgenomics.
DrugMetabDispos2011,39:2135–2147.
16.
WangQ,LiuQY,LiuZS,QianQ,SunQ,PanDY:InhibitionofhepatocelluarcarcinomaMAT2AandMAT2betageneexpressionsbysingleanddualsmallinterferingRNA.
JExpClinCancerRes2008,27:72–80.
17.
JaniTS,GobejishviliL,HotePT,BarveAS,Joshi-BarveS,KharebavaG,SuttlesJ,ChenT,McClainCJ,BarveS:InhibitionofmethionineadenosyltransferaseIIinducesFasLexpression,Fas-DISCformationandcaspase-8-dependentapoptoticdeathinTleukemiccells.
CellRes2009,19:358–369.
18.
AttiaRR,GardnerLA,MahrousE,TaxmanDJ,LegrosL,RoweS,TingJP,GellerA,KotbM:SelectivetargetingofleukemiccellgrowthinvivoandinvitrousingagenesilencingapproachtodiminishS-adenosylmethioninesynthesis.
JBiolChem2008,283:30788–30795.
19.
YangH,ZhengY,LiTW,PengH,FernandezRamosD,Martínez-ChantarML,RojasAL,MatoJM,LuSC:Methionineadenosyltransferase2B,HuRandsirtuin1crosstalkimpactsonResveratrol'seffectonapoptosisandgrowthinlivercancercells.
JBiolChem2013,288:23161–23170.
20.
Moran-JonesK,WaymanL,KennedyDD,ReddelRR,SaraS,SneeMJ,SmithR:HnRNPA2apotentialssDNA/RNAmolecularadapteratthetelomere.
NucleicAcidsRes2005,33:486–496.
21.
CarpenterB,MacKayC,AlnabulsiA,MacKayM,TelferC,MelvinWT,MurrayGI:Therolesofheterogeneousnuclearribonucleoproteinsintumourdevelopmentandprogression.
BiochimBiophysActa2006,1765:85–100.
22.
ChaudhuryA,ChanderP,HowePH:Heterogeneousnuclearribonucleoproteins(hnRNPs)incellularprocesses:FocusonhnRNPE1'smultifunctionalregulatoryroles.
RNA2010,16:1449–1462.
Review.
23.
Ostareck-LedererA,OstareckDH:Precisionmechanicswithmultifunctionaltools:howhnRNPKandhnRNPsE1/E2contributetopost-transcriptionalcontrolofgeneexpressioninhematopoiesis.
CurrProteinPeptSci2012,13:391–400.
24.
WangH,YeY,PanSY,ZhuGY,LiYW,FongDW,YuZL:ProteomicidentificationofproteinsinvolvedintheanticanceractivitiesoforidonininHepG2cells.
Phytomedicine2011,18:163–169.
25.
NiikuraY,OgiH,KikuchiK,KitagawaK:BUB3thatdissociatesfromBUB1activatescaspase-independentmitoticdeath(CIMD).
CellDeathDiffer2010,17:1011–1024.
26.
RobertsonC,ChurchSW,NagarHA,PriceJ,HallPA,RussellSE:PropertiesofSEPT9isoformsandtherequirementforGTPbinding.
JPathol2004,203:519–527.
27.
ChackoAD,McDadeSS,ChanduloyS,ChurchSW,KennedyR,PriceJ,HallPA,RussellSE:ExpressionoftheSEPT9_i4isoformconfersresistancetomicrotubule-interactingdrugs.
CellOncol2012,35:85–93.
28.
KimMS,FroeseCD,EsteyMP,TrimbleWS:SEPT9occupiestheterminalpositionsinseptinoctamersandmediatespolymerization-dependentfunctionsinabscission.
JCellBiol2011,195:815–826.
29.
CarvouN,HolicR,LiM,FutterC,SkippenA,CockcroftS:Phosphatidylinositol-andphosphatidylcholine-transferactivityofPITP-betaisessentialforCOPI-mediatedretrogradetransportfromtheGolgitotheendoplasmicreticulum.
JCellSci2010,123:1262–1273.
30.
SnoekGT:Phosphatidylinositoltransferproteins:emergingrolesincellproliferation,celldeathandsurvival.
IUBMBLife2004,56:467–475.
Review.
31.
RimanS,RizkallahR,KassardjianA,AlexanderKE,LüscherB,HurtMM:PhosphorylationofthetranscriptionfactorYY1byCK2αpreventscleavagebycaspase7duringapoptosis.
MolCellBiol2012,32:797–807.
32.
PayneKJ,DovatS:Ikarosandtumorsuppressioninacutelymphoblasticleukemia.
CritRevOncog2011,16:3–12.
Review.
33.
DovatS,SongC,PayneKJ,LiZ:Ikaros,CK2kinase,andtheroadtoleukemia.
MolCellBiochem2011,356:201–207.
Review.
34.
WienerR,ZhangX,WangT,WolbergerC:ThemechanismofOTUB1-mediatedinhibitionofubiquitination.
Nature2012,483:618–622.
35.
SunXX,ChallagundlaKB,DaiMS:Positiveregulationofp53stabilityandactivitybythedeubiquitinatingenzymeOtubain1.
EMBOJ2012,31:576–592.
36.
LudwigH,KhayatD,GiacconeG,FaconT:Proteasomeinhibitionanditsclinicalprospectsinthetreatmentofhematologicandsolidmalignancies.
Cancer2005,104:1794–1797.
37.
DavisCD,TsujiPA,MilnerJA:Selenoproteinsandcancerprevention.
AnnuRevNutr2012,32:73–95.
Review.
38.
CarletM,JanjetovicK,RainerJ,SchmidtS,Panzer-GrümayerR,MannG,PrelogM,MeisterB,PlonerC,KoflerR:Expression,regulationandfunctionofphosphofructo-kinase/fructose-biphosphatases(PFKFBs)inglucocorticoid-inducedapoptosisofacutelymphoblasticleukemiacells.
BMCCancer2010,10:638–649.
Costaetal.
ClinicalProteomics2014,11:31Page11of12http://www.
clinicalproteomicsjournal.
com/content/11/1/3139.
KowalskiW,NoconD,GamianA,KoodziejJ,RakusD:AssociationofC-terminalregionofphosphoglyceratemutasewithglycolyticcomplexregulatesenergyproductionincancercells.
JCellPhysiol2012,227:2613–2621.
40.
HullemanE,KazemierKM,HollemanA,VanderWeeleDJ,RudinCM,BroekhuisMJ,EvansWE,PietersR,DenBoerML:Inhibitionofglycolysismodulatesprednisoloneresistanceinacutelymphoblasticleukemiacells.
Blood2009,113:2014–2021.
41.
FootzTK,Brinkman-MillsP,BantingGS,MaierSA,RiaziMA,BridglandL,HuS,BirrenB,MinoshimaS,ShimizuN,PanH,NguyenT,FangF,FuY,RayL,WuH,ShaullS,PhanS,YaoZ,ChenF,HuanA,HuP,WangQ,LohP,QiS,RoeBA,McDermidHE:Analysisofthecateyesyndromecriticalregioninhumansandtheregionofconservedsyntenyinmice:asearchforcandidategenesatornearthehumanchromosome22pericentromere.
GenomeRes2001,11:1053–1070.
42.
MichaudJ,SimpsonKM,EscherR,Buchet-PoyauK,BeissbarthT,CarmichaelC,RitchieME,SchützF,CannonP,LiuM,ShenX,ItoY,RaskindWH,HorwitzMS,OsatoM,TurnerDR,SpeedTP,KavallarisM,SmythGK,ScottHS:IntegrativeanalysisofRUNX1downstreampathwaysandtargetgenes.
BMCGenomics2008,9:363–380.
43.
JiangN,KohGS,LimJY,KhamSK,AriffinH,ChewFT,YeohAE:BIMisaprognosticbiomarkerforearlyprednisoloneresponseinpediatricacutelymphoblasticleukemia.
ExpHematol2011,39:321–329.
44.
TrembleyJH,ChenZ,UngerG,SlatonJ,KrenBT,VanWaesC,AhmedK:EmergenceofproteinkinaseCK2asakeytargetincancertherapy.
Biofactors2010,36:187–195.
45.
PolakR,BuitenhuisM:ThePI3K/PKBsignalingmoduleaskeyregulatorofhematopoiesis:implicationsfortherapeuticstrategiesinleukemia.
Blood2012,119:911–923.
46.
ErnstA,AvvakumovG,TongJ,FanY,ZhaoY,AlbertsP,PersaudA,WalkerJR,NeculaiAM,NeculaiD,VorobyovA,GargP,BeattyL,ChanPK,JuangYC,LandryMC,YehC,ZeqirajE,KaramboulasK,Allali-HassaniA,VedadiM,TyersM,MoffatJ,SicheriF,PelletierL,DurocherD,RaughtB,RotinD,YangJ,MoranMF,etal:Astrategyformodulationofenzymesintheubiquitinsystem.
Science2013,339:590–595.
47.
SchneiderP,CostaO,LegrandE,BigotD,LecleireS,GrassiV,VannierJP,VasseM:Invitrosecretionofmatrixmetalloprotease9isaprognosticmarkerinchildhoodacutelymphoblasticleukemia.
LeukRes2010,34:24–31.
48.
GandemerV,ChevretS,PetitA,VermylenC,LeblancT,MichelG,SchmittC,LejarsO,SchneiderP,DemeocqF,Bader-MeunierB,BernaudinF,PerelY,AuclercMF,CayuelaJM,LevergerG,BaruchelA,FRALLEGroup:ExcellentprognosisoflaterelapsesofETV6/RUNX1-positivechildhoodacutelymphoblasticleukemia:lessonsfromtheFRALLE93protocol.
Haematologica2012,97:1743–1750.
49.
VanDongenJJ,MacintyreEA,GabertJA,DelabesseE,RossiV,SaglioG,GottardiE,RambaldiA,DottiG,GriesingerF,ParreiraA,GameiroP,DiázMG,MalecM,LangerakAW,SanMiguelJF,BiondiA:StandardizedRT-PCRanalysisoffusiongenetranscriptsfromchromosomeaberrationsinacuteleukemiafordetectionofminimalresidualdisease.
ReportoftheBIOMED-1ConcertedAction:investigationofminimalresidualdiseaseinacuteleukemia.
Leukemia1999,13:1901–1928.
50.
SchneiderP,VasseM,CorbièreC,LegrandE,Marie-CardineA,BoquetC,VannierJP:Endostatinvariationsinchildhoodacutelymphoblasticleukaemia-comparisonwithbasicfibroblastgrowthfactorandvascularendothelialgrowthfactor.
LeukRes2007,31:629–638.
51.
TnaniH,LópezI,JouenneT,VicientaCM:Proteincompositionanalysisofoilbodiesfrommaizeembryosduringgermination.
JPlantPhysiol2011,168:510–513.
52.
MagdeldinS,EnanyS,YoshidaY,XuB,ZhangY,ZureenaZ,LokamaniI,YaoitaE,YamamotoT:Basicsandrecentadvancesoftwodimensional-polyacrylamidegelelectrophoresis.
ClinProteomics2014,11:16–26.
doi:10.
1186/1559-0275-11-31Citethisarticleas:Costaetal.
:Proteomicprofileofpre-B2lymphoblastsfromchildrenwithacutelymphoblasticleukemia(ALL)inrelationwiththetranslocation(12;21).
ClinicalProteomics201411:31.
SubmityournextmanuscripttoBioMedCentralandtakefulladvantageof:ConvenientonlinesubmissionThoroughpeerreviewNospaceconstraintsorcolorgurechargesImmediatepublicationonacceptanceInclusioninPubMed,CAS,ScopusandGoogleScholarResearchwhichisfreelyavailableforredistributionSubmityourmanuscriptatwww.
biomedcentral.
com/submitCostaetal.
ClinicalProteomics2014,11:31Page12of12http://www.
clinicalproteomicsjournal.
com/content/11/1/31
- hnRNPswww.bu622.com相关文档
- uctswww.bu622.com
- wiewww.bu622.com
- 公司www.bu622.com
- 24.4www.bu622.com
- ERSEwww.bu622.com
- typicallywww.bu622.com
HostKvm($4.25/月),俄罗斯CN2带宽大升级,俄罗斯/香港高防限量5折优惠进行中
HostKvm是一家成立于2013年的国外VPS服务商,产品基于KVM架构,数据中心包括日本、新加坡、韩国、美国、俄罗斯、中国香港等多个地区机房,均为国内直连或优化线路,延迟较低,适合建站或者远程办公等。本月,商家旗下俄罗斯、新加坡、美国、香港等节点带宽进行了大幅度升级,俄罗斯机房国内电信/联通直连,CN2线路,150Mbps(原来30Mbps)带宽起,目前俄罗斯和香港高防节点5折骨折码继续优惠中...
RackNerd($199/月),5IP,1x256G SSD+2x3THDD
我们先普及一下常识吧,每年9月的第一个星期一是美国劳工节。于是,有一些服务商会基于这些节日推出吸引用户的促销活动,比如RackNerd有推出四款洛杉矶和犹他州独立服务器,1G带宽、5个独立IP地址,可以配置Windows和Linux系统,如果有需要独立服务器的可以看看。第一、劳工节促销套餐这里有提供2个套餐。两个方案是选择犹他州的,有2个方案是可以选择洛杉矶机房的。CPU内存SSD硬盘配置流量价格...
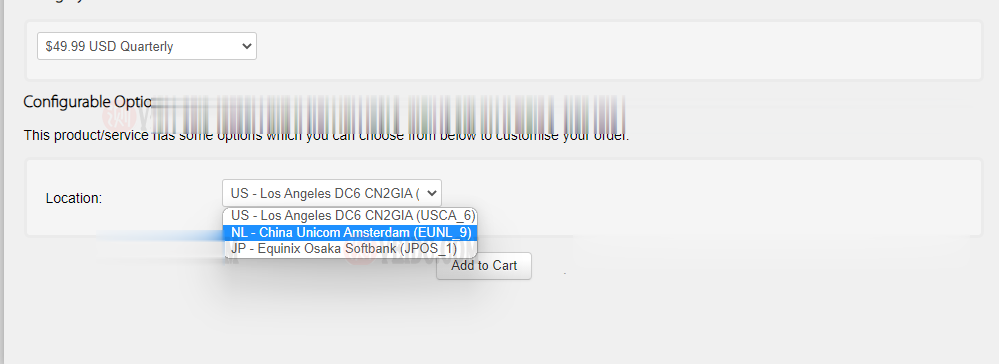
搬瓦工:新增荷兰机房 EUNL_9 测评,联通 AS10099/AS9929 高端优化路线/速度 延迟 路由 丢包测试
搬瓦工最近上线了一个新的荷兰机房,荷兰 EUNL_9 机房,这个 9 的编号感觉也挺随性的,之前的荷兰机房编号是 EUNL_3。这次荷兰新机房 EUNL_9 采用联通 AS9929 高端路线,三网都接入了 AS9929,对于联通用户来说是个好消息,又多了一个选择。对于其他用户可能还是 CN2 GIA 机房更合适一些。其实对于联通用户,这个荷兰机房也是比较远的,相比之下日本软银 JPOS_1 机房可...
www.bu622.com为你推荐
-
0.21网易yeah支付宝蜻蜓发布想做支付宝蜻蜓刷脸支付的代理么?怎么做?重庆电信断网电信光纤一直掉线,打电话问说是机房出了问题 要排查,已经一个星期了还没弄好,大概需要多久才能弄好?波音737起飞爆胎美国737MAX又紧急迫降,为什么它还在飞?重庆杨家坪猪肉摊主杀人重庆一市民发现买的新鲜猪肉晚上发蓝光.专家解释,猪肉中含磷较多且携带了一种能发光的细菌--磷光杆菌时flashfxp下载求最新无需注册的FlashFXP下载地址360arp防火墙在哪360ARP防火墙重庆电信dns重庆电信的DNS是什么邮件eset瑞东集团中粮集团主要生产什么的?是国企么