CHEK2ww.00271.com
ww.00271.com 时间:2021-03-23 阅读:()
Package'MoonFinder'October26,2019TypePackageTitleIdentifyMoonlightingncRNAsVersion1.
0.
3Date2019-10-1AuthorLixinChengMaintainerLixinChengDescriptionIdentifymoonlightingnon-codingRNAsatthegenomelevelbasedonthefunctionalan-notationandinteractomedataofncRNAsandproteins.
MoonFinderprovidesanextensiveframe-workforthedetectionofproteinmodulesandestablishmentofRNA-moduleassociations.
Importsannotate,igraph,org.
Hs.
eg.
db,pheatmap,clusterProler,RColorBrewer,grDevices,graphics,stats,utilsbiocViewsAnnotation,Clustering,GeneSetEnrichment,GO,MultipleComparisonVignetteBuilderknitrDependsR(>=3.
4)LicenseGPL-2RoxygenNote6.
1.
1Suggestsknitr,rmarkdown,usethisEncodingUTF-8NeedsCompilationnoRepositoryCRANDate/Publication2019-10-2610:30:02UTCRtopicsdocumented:MoonFinder-package2combinedModuleList3corumModuleList5moduleCombiding7moduleFunctionComparing812MoonFinder-packagemoduleHeatmap9moduleList10moduleMappingID12moonlightingCoefcient13pcdqModuleList14ppi16predModuleList16rna2mod18rna2module19rna2protein20SDI20simMatBP.
Jiang21simMatBP.
Lin22simMatBP.
Rel22simMatBP.
Resnik23simMatBP.
Wang23uniGene24Index25MoonFinder-packageMoonFinderDescriptionIdentifymoonlightingnon-codingRNAsDetailsMoonFinderisdevelopedfortheidenticationofmncRNAsatthegenomelevelbasedonthefunc-tionalannotationandinteractomedataofncRNAsandproteins.
MoonFinderprovidesanextensiveframeworkforthedetectionofproteinmodulesandestablishmentofRNA-moduleassociations.
Author(s)Maintainer:LixinChengReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examples##1)IdentifythemoonlightlncRNAs#install.
packages("ggraph")data("ppi","simMatBP.
Resnik","moduleList","rna2module")rnaStatMat=moonlightingCoefficient(rna2module,simMatBP.
Resnik)head(rnaStatMat)combinedModuleList3##2)SetuptheSDIthresholdaccordingtoSDIdistributionssdi=as.
numeric(rnaStatMat[,3])dev.
new(width=4,height=4)hist(sdi,border=FALSE,xlab="MC",xlim=c(0,1),ylim=c(0,4),breaks=seq(0,1,0.
02),prob=TRUE,main="Resnik")lines(density(sdi),lwd=2,col="orange")mlncRNA=rnaStatMat[sdi>0.
4,1]##3)Heatmap#IllustrationofthemediatedmodulesforagivenmoonlightingncRNA.
library(igraph)gReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesm1=c("Q15021","Q9BPX3","Q15003","O95347","Q9NTJ3")m2=c("Q92828","Q13227","O15379","O75376","O60907","Q9BZK7")m3=c("P61160","P61158","O15143","O15144","O15145","P59998","O15511")m4=c("Q92828","Q13227","O15379","O75376","Q15021")m5=c("P61160","P61158","O15143","O15144","O15145","Q9NTJ3")mList1=list(m1,m2,m3,m4,m5)mList2=moduleCombiding(mList1,OScutoff=0.
5)head(mList2)8moduleFunctionComparingmoduleFunctionComparingComparefunctionsoftargetmodulesDescriptionPlotaguretocomparethefunctionsenrichedbydifferentmodules.
UsagemoduleFunctionComparing(rna,rna2mod,modList)ArgumentsrnaanRNAsymbol.
rna2modNxM0-1binarymatrixofNRNAandMmodule.
modListalistofprotienmodules.
Valueadotplotcomparingthefunctionsenrichedbydistinctmodules.
Author(s)LixinChengReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Exampleslibrary(clusterProfiler)data(rna2module);data(moduleList)#moduleFunctionComparing("DIRC3",rna2module,moduleList)#moduleFunctionComparing("CRNDE",rna2module,moduleList)moduleHeatmap9moduleHeatmapmoduleHeatmapDescriptionDrawaheatmaptoillustratetheproteinmodulesmediatedbyanRNA.
UsagemoduleHeatmap(rna,rna2mod,modList,net)ArgumentsrnaRNAsymbol.
rna2modNxM0-1binarymatrixofNRNAandMmodule.
modListalistofprotienmoudles.
netaprotein-proteininteractionnetworkcompiledinigraph.
ValueaheatmapgureindicatingproteinmodulestargetedbyanRNA.
Author(s)LixinChengReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Exampleslibrary(igraph)data(ppi)data(rna2module)data(moduleList)gReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesm1=c("Q15021","Q9BPX3","Q15003","O95347","Q9NTJ3")m2=c("Q92828","Q13227","O15379","O75376","O60907","Q9BZK7")m3=c("P61160","P61158","O15143","O15144","O15145","P59998","O15511")uniprotModuleList=list(m1,m2,m3)symbolModuleList=moduleMappingID(uniprotModuleList)names(symbolModuleList)=paste0("M",1:length(symbolModuleList))head(symbolModuleList)moonlightingCoefcient13moonlightingCoefficientmoonlightingCoefcientDescriptionCalculatethemoonlightingcoefcientforRNAs.
UsagemoonlightingCoefficient(rna2mod,simMat)Argumentsrna2modtwo-columnmatrixwitheachrowshowsthebinaryassociationbetweenRNAandprotein.
simMatadjacentmatrixindicatingthesemanticsimilarityamongproteinmodulesValuernaStatatableincludingncRNAsymble,numberoftargetmodules,andcorespondingmoonlightingcoefcient.
Author(s)LixinChengReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(rna2module)data(simMatBP.
Rel)mcOutput=moonlightingCoefficient(rna2module,simMat=simMatBP.
Rel)head(mcOutput)14pcdqModuleListpcdqModuleListpcdqModuleListDescriptionalistofmodulescollectedfromdatabasePCDq.
Usagedata("pcdqModuleList")FormatTheformatis:Listof412$pcdq_1:chr[1:39]"TPP1""A2M""GBA""GAPDH".
.
.
$pcdq_2:chr[1:41]"IKBKB""CHUK""MAP3K7""TUBB4A".
.
.
$pcdq_3:chr[1:24]"MED7""MED7""MED14""MED6".
.
.
$pcdq_4:chr[1:12]"BRCA1""MYC""ESR1""TP53".
.
.
$pcdq_5:chr[1:23]"PIK3R2""ABL1""EGFR""EGF".
.
.
$pcdq_6:chr[1:19]"FOS""FOS""JUN""FOSL1".
.
.
$pcdq_7:chr[1:14]"SF3B1""CD2BP2""U2AF2""YWHAG".
.
.
$pcdq_8:chr[1:10]"KAT7""VIM""SAT1""CHD3".
.
.
$pcdq_9:chr[1:6]"RTCA""HLA-B""IMPDH2""MCM3".
.
.
$pcdq_10:chr[1:18]"ZNHIT1""YEATS4""ACTL6A""MORF4L2".
.
.
$pcdq_11:chr[1:23]"PSMA7""EIF1B""EIF1B""EPB41".
.
.
$pcdq_12:chr[1:14]"PSMD11""SMAD7""SMAD9""SMAD6".
.
.
$pcdq_13:chr[1:13]"NCK1""SFPQ""SFPQ""PDIA3".
.
.
$pcdq_14:chr[1:17]"ORC5""ORC5""ORC4""CDK1".
.
.
$pcdq_15:chr[1:7]"APOA1""TTR""ALB""TF".
.
.
$pcdq_16:chr[1:7]"HGS""STAM2""DAZAP2""UBQLN4".
.
.
$pcdq_17:chr[1:6]"LDOC1""HNRNPC""FXR2""CCDC85B".
.
.
$pcdq_18:chr[1:7]"PLSCR1""SPRY2""GLRX3""EFEMP2".
.
.
$pcdq_19:chr[1:7]"BCL2""BID""BAX""BCL2L1".
.
.
$pcdq_20:chr[1:7]"GH1""RPL3""RPL3""EIF6".
.
.
$pcdq_21:chr[1:11]"CFLAR""FAS""FASLG""FASLG".
.
.
$pcdq_22:chr[1:10]"PIAS1""TP53""AR""SUMO1".
.
.
$pcdq_23:chr[1:12]"RB1""MYBL1""MYBL2""RBL2".
.
.
$pcdq_24:chr[1:8]"FYB1""FYN""HCK""SRC".
.
.
$pcdq_25:chr[1:12]"TNF""TNF""TNFRSF1A""TNFRSF1B".
.
.
$pcdq_26:chr[1:8]"FAM107A""FAM107A""KRT19""KRT15".
.
.
$pcdq_27:chr[1:10]"RBFOX2""CCNK""ATXN1""DAZAP2".
.
.
$pcdq_28:chr[1:13]"WDR5""RBX1""TLE2""CUL4A".
.
.
$pcdq_29:chr[1:24]"PSMD11""PSMD12""PSMD14""PSMD3".
.
.
$pcdq_30:chr[1:5]"SPTBN1""SP-TAN1""CEP63""EXOC1".
.
.
$pcdq_31:chr[1:20]"GEMIN2""SNRPB""SNRPB""GEMIN4".
.
.
$pcdq_32:chr[1:11]"ORC4""MCM3""MCM4""MCM5".
.
.
$pcdq_33:chr[1:16]"DXO""ZFP36""EXOSC10""EXOSC9".
.
.
$pcdq_34:chr[1:6]"CTNND1""CDH1""CTNNA1""CTNNB1".
.
.
$pcdq_35:chr[1:11]"RAD21""MYC""SMC1A""PDS5A".
.
.
$pcdq_36:chr[1:9]"CDC42""RAC1""YWHAH""PRKCZ".
.
.
$pcdq_37:chr[1:6]"RFXANK""HN-RNPM""C20orf24""NUP93".
.
.
$pcdq_38:chr[1:7]"MCC""VHL""ELOC""CUL2".
.
.
$pcdq_39:chr[1:14]"POLR2D""POLR2C""POLR2C""POLR2E".
.
.
$pcdq_40:chr[1:11]"BRCA1""NBN""CHEK2""H2AFX".
.
.
$pcdq_41:chr[1:8]"NUP214""NUP62""NUP153""RANBP2".
.
.
$pcdq_42:chr[1:8]"IFNAR1""TYK2""IL6ST""IL6ST".
.
.
$pcdq_43:chr[1:18]"ARID1A""ACTL6A""SMARCA2""SMARCA4".
.
.
$pcdq_44:chr[1:64]"PRMT5""RAD51B""HIST1H2BK""HIST1H2BJ".
.
.
$pcdq_45:chr[1:9]"HRAS""RAF1""BRAF""MAPK1".
.
.
$pcdq_46:chr[1:7]"TUBG1""AKT1""AKT2""GSK3A".
.
.
$pcdq_47:chr[1:8]"THRB""VDR""RXRA""PRMT2".
.
.
$pcdq_48:chr[1:8]"MEN1""RPA2""RPA1""RPA3".
.
.
$pcdq_49:chr[1:8]"APAF1""CASP1""CASP3""CASP9".
.
.
$pcdq_50:chr[1:9]"BACH1"pcdqModuleList15"CREB3""CREB1""CREB1".
.
.
$pcdq_51:chr[1:7]"TXNDC9""PTN""CSTF2""BAG6".
.
.
$pcdq_52:chr[1:20]"SAP18""HDAC3""NCOR1""SAP30".
.
.
$pcdq_53:chr[1:5]"GFI1B""ACTN1""TRIM27""MAGEA11".
.
.
$pcdq_54:chr[1:13]"ARPC1B""ARPC2""ARPC2""ARPC3".
.
.
$pcdq_55:chr[1:7]"KPNA3""ARRB2""ARRB2""CSNK2A1".
.
.
$pcdq_56:chr[1:7]"TP53""HTT""NAP1L1""CTBP1".
.
.
$pcdq_57:chr[1:9]"EIF4G3""EIF4E""EIF4E""EIF4A2".
.
.
$pcdq_58:chr[1:10]"SNRNP200""MYC""HNRNPC""TOP1".
.
.
$pcdq_59:chr[1:9]"FANCG""FANCG""FANCA""FANCC".
.
.
$pcdq_60:chr[1:7]"DVL2""ZNF250""FXR2""AP2M1".
.
.
$pcdq_61:chr[1:6]"SNAPIN""BLOC1S1""BLOC1S4""BLOC1S2".
.
.
$pcdq_62:chr[1:8]"DDX6""ZFP36""DCP1A""EDC4".
.
.
$pcdq_63:chr[1:7]"GDF9""ZNF24""APLP1""EEF1A1".
.
.
$pcdq_64:chr[1:13]"IRAK2""IL1R1""IRAK1""TIRAP".
.
.
$pcdq_65:chr[1:10]"COPS2""GPS1""COPS4""COPS3".
.
.
$pcdq_66:chr[1:6]"SMC2""NCAPH""NCAPD2""NEK6".
.
.
$pcdq_67:chr[1:7]"WWP2""EWSR1""HERPUD1""HER-PUD1".
.
.
$pcdq_68:chr[1:14]"PPP2R1A""PPP2R1B""PPP2CB""PPP2R2A".
.
.
$pcdq_69:chr[1:5]"GAS7""NCKAP1""ABI1""CYFIP2".
.
.
$pcdq_70:chr[1:9]"CBL""CBLB""PDCD6IP""SH3KBP1".
.
.
$pcdq_71:chr[1:9]"CDK4""CCND1""CCND1""CCND3".
.
.
$pcdq_72:chr[1:11]"ACTL6A""TCF3""RUVBL2""INO80C".
.
.
$pcdq_73:chr[1:10]"LSM1""LSM2""LSM8""SNRPE".
.
.
$pcdq_74:chr[1:25]"SF3B1""SNRPB""SNRPB""U2AF2".
.
.
$pcdq_75:chr[1:5]"BCR""CFTR""COPB1""PDZK1".
.
.
$pcdq_76:chr[1:7]"TMSB4X""LIMS1""ERH""ILK".
.
.
$pcdq_77:chr[1:6]"GNAI2""FLNA""CANX""MTNR1A".
.
.
$pcdq_78:chr[1:6]"TPD52L2""TPD52L2""TPD52""TPD52L1".
.
.
$pcdq_79:chr[1:9]"KRT6A""KRT6C""KRT18""KRT8".
.
.
$pcdq_80:chr[1:7]"ERCC2""ERCC3""ERCC5""GTF2H1".
.
.
$pcdq_81:chr[1:5]"DNMT1""DNMT1""SUZ12""EZH2".
.
.
$pcdq_82:chr[1:9]"TGFB1""TGFB3""ENG""ENG".
.
.
$pcdq_83:chr[1:6]"CRYAA""CRYAA2""CRYAB""HSPB1".
.
.
$pcdq_84:chr[1:6]"MAFG""MAFK""JUN""NFE2L2".
.
.
$pcdq_85:chr[1:20]"ITGB3""ITGB1""ITGAV""ITGA5".
.
.
$pcdq_86:chr[1:8]"PARP1""XRCC6""XRCC5""XRCC1".
.
.
$pcdq_87:chr[1:7]"CDC37""CCT7""TANK""TANK".
.
.
$pcdq_88:chr[1:9]"CBX4""E2F6""BMI1""COMMD3-BMI1".
.
.
$pcdq_89:chr[1:8]"CDK11B""CDK11A""YWHAQ""YWHAB".
.
.
$pcdq_90:chr[1:6]"PEX14""ABCD3""ABCD1""PEX19".
.
.
$pcdq_91:chr[1:10]"POP7""RPP14""POP4""RPP38".
.
.
$pcdq_92:chr[1:7]"UBE2M""NEDD8""CAV1""CAV1".
.
.
$pcdq_93:chr[1:6]"F2""VTN""ITGB3""SERPINE1".
.
.
$pcdq_94:chr[1:5]"CD4""NR3C1""LCK""HSP90AA1".
.
.
$pcdq_95:chr[1:10]"NDC80""ZWINT""CBX5""CBX3".
.
.
$pcdq_96:chr[1:6]"ASB1""ASB6""ASB12""CUL5".
.
.
$pcdq_97:chr[1:7]"MYC""PCNA""RFC4""RFC2".
.
.
$pcdq_98:chr[1:7]"MEN1""WDR5""KMT2A""RBBP5".
.
.
$pcdq_99:chr[1:6]"TOM1L1""PTBP1""HNRNPH1""CRMP1".
.
.
[listoutputtruncated]ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(pcdqModuleList)head(pcdqModuleList)16predModuleListppippiDescriptiontwo-colomnmatrixwitheachrowshowsinteractionbetweenapairofproteins.
Usagedata("ppi")FormatTheformatis:chr[1:210410,1:2]"KANSL1L""SLC12A8""RBM47""RBM47""RBM47""RBM47""RBM47""RBM47""TTC26""TTC26""TTC26".
.
.
-attr(*,"dimnames")=Listof2.
.
$:NULL.
.
$:chr[1:2]"V1""V2"ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(ppi)head(ppi)predModuleListpredictedmodulesDescriptionalistofproteinmoudlesidentiedusingClusterONE.
Usagedata("predModuleList")FormatTheformatis:Listof837$pred_1:chr[1:216]"FBXO4""FBXL3""FBXW2""ISY1".
.
.
$pred_2:chr[1:169]"TNRC6A""SH3RF2""GPX8""SAP25".
.
.
$pred_3:chr[1:17]"NXT1""NUDT5""IKZF2""FBXO5".
.
.
$pred_4:chr[1:228]"PDK3""MAPRE1""TSC22D1""ELF2".
.
.
$pred_5:chr[1:51]"POLR2F""PSME3""MAGOH""VBP1".
.
.
$pred_6:chr[1:17]"FAU""RPL23""UBE2D2""RPS15".
.
.
$pred_7:chr[1:559]"RPL36AL""L3MBTL2""MUL1""RNF166".
.
.
$pred_8:chr[1:22]"NUBP1""RAD23A""RAD23B""EIF5".
.
.
$pred_9:chr[1:60]"MAK16""NIFK""GTPBP4""WDR33".
.
.
$pred_10:chr[1:296]"DYRK3""NXNL2"predModuleList17"ANKS6""NAIF1".
.
.
$pred_11:chr[1:10]"EHBP1""GPKOW""SNAPC3""CLP1".
.
.
$pred_12:chr[1:144]"ZNF165""PAPOLA""BRCA2""PSMD7".
.
.
$pred_13:chr[1:46]"DAAM2""PRPF39""RPAIN""KTN1".
.
.
$pred_14:chr[1:863]"FANCL""RBFOX1""BABAM1""IRAK4".
.
.
$pred_15:chr[1:80]"TRA2B""UBE2B""PPP2R2A""SNRPN".
.
.
$pred_16:chr[1:77]"LMX1A""TOPBP1""ZNF516""DHX38".
.
.
$pred_17:chr[1:63]"ZNF587B""NACA""PRPF4B""HDAC1".
.
.
$pred_18:chr[1:253]"ELP5""LMX1A""SH3RF2""GPX8".
.
.
$pred_19:chr[1:45]"RELB""GALK2""AMPD2""CTBS".
.
.
$pred_20:chr[1:6]"SMG9""NAPB""UPF3A""WNT8A".
.
.
$pred_21:chr[1:69]"ZNF45""ZNF33A""ZNF33B""PPARA".
.
.
$pred_22:chr[1:430]"NOL8""DHX30""EIF3M""MICAL3".
.
.
$pred_23:chr[1:186]"NPAT""EIF2B1""EIF4A2""TCEB3".
.
.
$pred_24:chr[1:315]"MACROD1""PAAF1""RIOK1""LARP6".
.
.
$pred_25:chr[1:6]"DDX18""SDAD1""NLE1""RBM28".
.
.
$pred_26:chr[1:56]"HNRNPH3""HNRNPH1""STIP1""ARRB2".
.
.
$pred_27:chr[1:43]"WDR1""TDO2""POLD2""ALDH9A1".
.
.
$pred_28:chr[1:14]"SLC25A16""GLB1""PPP3CB""PFKL".
.
.
$pred_29:chr[1:32]"EXOSC4""ISYNA1""NOX4""DCP1A".
.
.
$pred_30:chr[1:296]"NACA""PRPF4B""HDAC1""TDG".
.
.
$pred_31:chr[1:199]"TSC22D1""NAB2""TAB1""MAPK11".
.
.
$pred_32:chr[1:33]"LTA4H""HSPA1A""HSPA1B""U2AF1L5".
.
.
$pred_33:chr[1:6]"RBMX""IFNGR2""EIF4A3""IGHMBP2".
.
.
$pred_34:chr[1:37]"CCT8""CCT4""ANXA11""PAPOLA".
.
.
$pred_35:chr[1:12]"GLTSCR1L""ZMAT3""CWC22""XAB2".
.
.
$pred_36:chr[1:183]"TMEM62""FAAP100""CPSF1""KIF1A".
.
.
$pred_37:chr[1:99]"HBG2""PSPH""RBM6""RPP30".
.
.
$pred_38:chr[1:214]"SSBP2""ERH""RPL19""SRSF3".
.
.
$pred_39:chr[1:60]"USF1""ME2""GLDC""WARS".
.
.
$pred_40:chr[1:17]"NAT10""DDX47""ESF1""HEATR1".
.
.
$pred_41:chr[1:134]"RNH1""RARG""ANXA8""ENO3".
.
.
$pred_42:chr[1:13]"ZNF251""ZNF2""ZNF426""ZNF416".
.
.
$pred_43:chr[1:25]"METTL25""DDX51""KRI1""SRFBP1".
.
.
$pred_44:chr[1:21]"EIF4A1""RPS20""PRPS1""SHFM1".
.
.
$pred_45:chr[1:23]"SMC4""MAN2C1""SACM1L""OLA1".
.
.
$pred_46:chr[1:55]"BRPF1""EIF6""SIK1""WDR4".
.
.
$pred_47:chr[1:62]"PDHA2""BLVRB""PRDX5""PEBP1".
.
.
$pred_48:chr[1:24]"CNGB1""GAPVD1""HERPUD1""LAPTM4A".
.
.
$pred_49:chr[1:31]"TRIM2""FAM117A""INO80B""CCSER2".
.
.
$pred_50:chr[1:7]"RBM22""CWC25""CWC15""CPSF2".
.
.
$pred_51:chr[1:6]"ZNF160""VTA1""ZNF331""CELSR1".
.
.
$pred_52:chr[1:8]"ZNF324B""ZNF470""ZNF425""ZNF529".
.
.
$pred_53:chr[1:9]"ZNF225""ZNF221""ZNF510""ZNF256".
.
.
$pred_54:chr[1:65]"ISYNA1""NOX4""RIC8A""RRAGD".
.
.
$pred_55:chr[1:37]"SSR1""NOP2""NSF""MAP2K3".
.
.
$pred_56:chr[1:29]"PSMB10""RBM34""IL12RB1""CDKN2B".
.
.
$pred_57:chr[1:58]"POM121C""PODXL""KIF3C""NRP1".
.
.
$pred_58:chr[1:26]"WDR61""INTS2""NELFA""TAF1D".
.
.
$pred_59:chr[1:20]"GNL1""LTBR""GPX4""HSD17B3".
.
.
$pred_60:chr[1:14]"RFC4""RFC2""RFC1""GTF2F1".
.
.
$pred_61:chr[1:14]"U2AF1L4""PPIL4""CCDC12""LEO1".
.
.
$pred_62:chr[1:8]"MRPS28""MRPS7""MRPS2""COQ4".
.
.
$pred_63:chr[1:25]"SUPT20H""ARMC2""NCOA7""SLC30A5".
.
.
$pred_64:chr[1:26]"MAPK3""MARK3""RPA1""APEX1".
.
.
$pred_65:chr[1:27]"MAST3""RASAL1""SNAPIN""PAK4".
.
.
$pred_66:chr[1:7]"STBD1""FEN1""MLH1""IL15".
.
.
$pred_67:chr[1:34]"STMN1""YBX3""ZFX""RHOQ".
.
.
$pred_68:chr[1:23]"RINT1""BZW1""NOXA1""LAPTM4B".
.
.
$pred_69:chr[1:61]"RAMP1""BCAS2""DNAJC8""SMNDC1".
.
.
$pred_70:chr[1:121]"MLXIPL""ISYNA1""NOX4""DCP1A".
.
.
$pred_71:chr[1:49]"MAN1A1""SPR""CBS""GCK".
.
.
$pred_72:chr[1:14]"ZNF721""ZFP90""ZNF473""ZNF480".
.
.
$pred_73:chr[1:16]"ZNF433""ZNF283""CNTD1""ZNF614".
.
.
$pred_74:chr[1:69]"RBM25""EIF2B2""HINT1""STK19".
.
.
$pred_75:chr[1:24]"SULT1C4""EPRS""HSP90AA1""GALT".
.
.
$pred_76:chr[1:8]"AHI1""HUS1B""ALMS1""CCHCR1".
.
.
$pred_77:chr[1:75]"ALB""MAP4K3""TBC1D2""NUF2".
.
.
$pred_78:chr[1:37]"C11orf49""TMEM231""C14orf93""ZMAT3".
.
.
$pred_7918rna2mod:chr[1:21]"CFB""LHB""PRL""GCG".
.
.
$pred_80:chr[1:19]"GIPC3""GIPC2""BTBD6""LRR1".
.
.
$pred_81:chr[1:12]"ZNF667""ZNF484""ZNF569""ZNF684".
.
.
$pred_82:chr[1:23]"MRPS5""MRPS36""MRPS11""MRPS15".
.
.
$pred_83:chr[1:14]"DDX47""ESF1""HEATR1""PIF1".
.
.
$pred_84:chr[1:12]"C14orf93""FBRS""GPBP1L1""ZNF532".
.
.
$pred_85:chr[1:11]"ZNF605""ZNF554""ZNF429""ZNF677".
.
.
$pred_86:chr[1:9]"ZNF251""ZNF2""ZNF426""EFCAB11".
.
.
$pred_87:chr[1:12]"RMI1""CLSPN""FANCE""UBE2T".
.
.
$pred_88:chr[1:40]"LGALSL""PARP14""PLEKHN1""ZC2HC1C".
.
.
$pred_89:chr[1:48]"TSPAN2""TSPAN3""SNAP91""RAMP2".
.
.
$pred_90:chr[1:8]"NUP37""NUP35""CASC5""FBXO30".
.
.
$pred_91:chr[1:12]"APOL2""LBHD1""ACBD6""ZFP14".
.
.
$pred_92:chr[1:29]"SMG9""NAPB""UPF3A""WNT8A".
.
.
$pred_93:chr[1:33]"EIF4E""ENO1""PYGL""GPI".
.
.
$pred_94:chr[1:11]"XDH""RFX3""PIP4K2A""SOX2".
.
.
$pred_95:chr[1:26]"COX20""RBM48""CELF3""SZT2".
.
.
$pred_96:chr[1:14]"ZNF613""ZNF790""ZNF562""ZNF782".
.
.
$pred_97:chr[1:8]"SNAPC4""CPSF3L""RPRD2""TAF3".
.
.
$pred_98:chr[1:24]"ZNF821""DAB1""PLCG2""KLRC2".
.
.
$pred_99:chr[1:15]"RANGAP1""NUP153""RANBP2""GSK3A".
.
.
[listoutputtruncated]ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(predModuleList)head(predModuleList)rna2modrna2modDescriptionConstructa0-1binarymatrixtorepresenttheassociationbetweenRNAsandproteinmodules.
Usagerna2mod(rna2prot,modlist,pCutoff=0.
01,bgProtNum)Argumentsrna2prottwo-columnmatrixwitheachrowshowsthebinaryassociationbetweenRNAandprotein.
modlistalistofproteinmodules.
pCutoffthresholdoftheP-valueforenrichmentanalysis.
bgProtNumthenumberofbackgroundproteins.
Valuerna2moduleNxM0-1binarymatrixofNRNAandMmodule.
moduleListalistofmodulestargetedbyatleastoneRNA.
rna2module19Author(s)LixinChengReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(rna2protein)data(combinedModuleList)data(uniGene)uniGeneNum=length(uniGene)result=rna2mod(rna2protein[1:100,],combinedModuleList,pCutoff=0.
01,uniGeneNum)rna2module=result[[1]]moduleList=result[[2]]rna2moduleRNA-moduleassociationmatrixDescriptionA0-1binarymatrixrepresentingtheassociationbetweenRNAsandmodulesUsagedata("rna2module")FormatTheformatis:num[1:776,1:175]0000000000.
.
.
-attr(*,"dimnames")=Listof2.
.
$:chr[1:776]"CRNDE""NEAT1""ZFAS1""GAS5"chr[1:175]"1""2""3""4".
.
.
ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(rna2module)head(rna2module)20SDIrna2proteinrna2proteinDescriptiontwo-columnmatrixwitheachrowshowsthebinaryassociationbetweenRNAandprotein.
Usagedata("rna2protein")FormatTheformatis:chr[1:12008,1:2]"LINC00271""MALAT1""CRNDE""PINK1-AS""TUG1""MALAT1""CRNDE""C11orf95""ANCR""LOC647323".
.
.
-attr(*,"dimnames")=Listof2.
.
$:NULL.
.
$:chr[1:2]"Interactor1""Interactor2"ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(rna2protein)head(rna2protein)SDIShannondiversityindexDescriptionCalculatetheSimpson'sdiversityindex(SDI).
UsageSDI(x)Argumentsxweightsoflatentfeatures.
Valuezmoonlightingcoefcient,ornormalizedShannon'sdiversityindex.
simMatBP.
Jiang21Author(s)LixinChengReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesa=sample(1:10,4)b=a/sum(a)SDI(b)simMatBP.
Jiangasemanticsimilarity(Jiang)matrixDescriptionAnadjacentmatrixofthesemanticsimilarity(Jiang)amongdiffernetproteinmodules.
Usagedata("simMatBP.
Jiang")FormatTheformatis:num[1:175,1:175]10.
8570.
9350.
8680.
5470.
780.
7130.
7750.
7790.
807.
.
.
-attr(*,"dimnames")=Listof2.
.
$:chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4"chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4".
.
.
ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(simMatBP.
Jiang)dim(simMatBP.
Jiang)22simMatBP.
RelsimMatBP.
Linasemanticsimilarity(Lin)matrixDescriptionAnadjacentmatrixofthesemanticsimilarity(Lin)amongdiffernetproteinmodules.
Usagedata("simMatBP.
Lin")FormatTheformatis:num[1:175,1:175]10.
8770.
9450.
8860.
5610.
8080.
7330.
7930.
8050.
825.
.
.
-attr(*,"dimnames")=Listof2.
.
$:chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4"chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4".
.
.
ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(simMatBP.
Lin)head(simMatBP.
Lin)simMatBP.
Relasemanticsimilarity(Rel)matrixDescriptionAnadjacentmatrixofthesemanticsimilarity(Rel)amongdiffernetproteinmodules.
Usagedata("simMatBP.
Rel")FormatTheformatis:num[1:175,1:175]0.
9940.
870.
9390.
8790.
5340.
7980.
720.
7840.
7950.
815.
.
.
-attr(*,"dimnames")=Listof2.
.
$:chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4"chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4".
.
.
ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
simMatBP.
Resnik23Examplesdata(simMatBP.
Rel)head(simMatBP.
Rel)simMatBP.
Resnikasemanticsimilarity(Resnik)matrixDescriptionAnadjacentmatrixofthesemanticsimilarity(Resnik)amongdiffernetproteinmodules.
Usagedata("simMatBP.
Resnik")FormatTheformatis:num[1:175,1:175]0.
5860.
4980.
5420.
5050.
2960.
4550.
4010.
4420.
4510.
46.
.
.
-attr(*,"dimnames")=Listof2.
.
$:chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4"chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4".
.
.
ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(simMatBP.
Resnik)head(simMatBP.
Resnik)simMatBP.
Wangasemanticsimilarity(Wang)matrix~DescriptionAnadjacentmatrixofthesemanticsimilarity(Wang)amongdiffernetproteinmodules.
Usagedata("simMatBP.
Wang")FormatTheformatis:num[1:175,1:175]10.
8270.
9190.
8450.
5270.
7610.
6590.
7250.
7420.
77.
.
.
-attr(*,"dimnames")=Listof2.
.
$:chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4"chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4".
.
.
24uniGeneReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(simMatBP.
Wang)head(simMatBP.
Wang)uniGeneBackgroundgenesymbolsDescriptionAllthegenesymbolsusedintheentireprotieninteractionnetwork.
Usagedata("uniGene")FormatTheformatis:chr[1:10111]"A1CF""AAAS""AADAT""AAGAB""AAK1""AAMDC""AAMP""AAR2""AARS""AARS2""AARSD1""AASDHPPT".
.
.
ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(uniGene)head(uniGene)IndexTopicdatasetscombinedModuleList,3corumModuleList,5moduleList,10pcdqModuleList,14ppi,16predModuleList,16rna2module,19rna2protein,20simMatBP.
Jiang,21simMatBP.
Lin,22simMatBP.
Rel,22simMatBP.
Resnik,23simMatBP.
Wang,23uniGene,24TopicpackageMoonFinder-package,2combinedModuleList,3corumModuleList,5moduleCombiding,7moduleFunctionComparing,8moduleHeatmap,9moduleList,10moduleMappingID,12MoonFinder(MoonFinder-package),2MoonFinder-package,2moonlightingCoefficient,13pcdqModuleList,14ppi,16predModuleList,16rna2mod,18rna2module,19rna2protein,20SDI,20simMatBP.
Jiang,21simMatBP.
Lin,22simMatBP.
Rel,22simMatBP.
Resnik,23simMatBP.
Wang,23uniGene,2425
0.
3Date2019-10-1AuthorLixinChengMaintainerLixinChengDescriptionIdentifymoonlightingnon-codingRNAsatthegenomelevelbasedonthefunctionalan-notationandinteractomedataofncRNAsandproteins.
MoonFinderprovidesanextensiveframe-workforthedetectionofproteinmodulesandestablishmentofRNA-moduleassociations.
Importsannotate,igraph,org.
Hs.
eg.
db,pheatmap,clusterProler,RColorBrewer,grDevices,graphics,stats,utilsbiocViewsAnnotation,Clustering,GeneSetEnrichment,GO,MultipleComparisonVignetteBuilderknitrDependsR(>=3.
4)LicenseGPL-2RoxygenNote6.
1.
1Suggestsknitr,rmarkdown,usethisEncodingUTF-8NeedsCompilationnoRepositoryCRANDate/Publication2019-10-2610:30:02UTCRtopicsdocumented:MoonFinder-package2combinedModuleList3corumModuleList5moduleCombiding7moduleFunctionComparing812MoonFinder-packagemoduleHeatmap9moduleList10moduleMappingID12moonlightingCoefcient13pcdqModuleList14ppi16predModuleList16rna2mod18rna2module19rna2protein20SDI20simMatBP.
Jiang21simMatBP.
Lin22simMatBP.
Rel22simMatBP.
Resnik23simMatBP.
Wang23uniGene24Index25MoonFinder-packageMoonFinderDescriptionIdentifymoonlightingnon-codingRNAsDetailsMoonFinderisdevelopedfortheidenticationofmncRNAsatthegenomelevelbasedonthefunc-tionalannotationandinteractomedataofncRNAsandproteins.
MoonFinderprovidesanextensiveframeworkforthedetectionofproteinmodulesandestablishmentofRNA-moduleassociations.
Author(s)Maintainer:LixinChengReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examples##1)IdentifythemoonlightlncRNAs#install.
packages("ggraph")data("ppi","simMatBP.
Resnik","moduleList","rna2module")rnaStatMat=moonlightingCoefficient(rna2module,simMatBP.
Resnik)head(rnaStatMat)combinedModuleList3##2)SetuptheSDIthresholdaccordingtoSDIdistributionssdi=as.
numeric(rnaStatMat[,3])dev.
new(width=4,height=4)hist(sdi,border=FALSE,xlab="MC",xlim=c(0,1),ylim=c(0,4),breaks=seq(0,1,0.
02),prob=TRUE,main="Resnik")lines(density(sdi),lwd=2,col="orange")mlncRNA=rnaStatMat[sdi>0.
4,1]##3)Heatmap#IllustrationofthemediatedmodulesforagivenmoonlightingncRNA.
library(igraph)gReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesm1=c("Q15021","Q9BPX3","Q15003","O95347","Q9NTJ3")m2=c("Q92828","Q13227","O15379","O75376","O60907","Q9BZK7")m3=c("P61160","P61158","O15143","O15144","O15145","P59998","O15511")m4=c("Q92828","Q13227","O15379","O75376","Q15021")m5=c("P61160","P61158","O15143","O15144","O15145","Q9NTJ3")mList1=list(m1,m2,m3,m4,m5)mList2=moduleCombiding(mList1,OScutoff=0.
5)head(mList2)8moduleFunctionComparingmoduleFunctionComparingComparefunctionsoftargetmodulesDescriptionPlotaguretocomparethefunctionsenrichedbydifferentmodules.
UsagemoduleFunctionComparing(rna,rna2mod,modList)ArgumentsrnaanRNAsymbol.
rna2modNxM0-1binarymatrixofNRNAandMmodule.
modListalistofprotienmodules.
Valueadotplotcomparingthefunctionsenrichedbydistinctmodules.
Author(s)LixinChengReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Exampleslibrary(clusterProfiler)data(rna2module);data(moduleList)#moduleFunctionComparing("DIRC3",rna2module,moduleList)#moduleFunctionComparing("CRNDE",rna2module,moduleList)moduleHeatmap9moduleHeatmapmoduleHeatmapDescriptionDrawaheatmaptoillustratetheproteinmodulesmediatedbyanRNA.
UsagemoduleHeatmap(rna,rna2mod,modList,net)ArgumentsrnaRNAsymbol.
rna2modNxM0-1binarymatrixofNRNAandMmodule.
modListalistofprotienmoudles.
netaprotein-proteininteractionnetworkcompiledinigraph.
ValueaheatmapgureindicatingproteinmodulestargetedbyanRNA.
Author(s)LixinChengReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Exampleslibrary(igraph)data(ppi)data(rna2module)data(moduleList)gReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesm1=c("Q15021","Q9BPX3","Q15003","O95347","Q9NTJ3")m2=c("Q92828","Q13227","O15379","O75376","O60907","Q9BZK7")m3=c("P61160","P61158","O15143","O15144","O15145","P59998","O15511")uniprotModuleList=list(m1,m2,m3)symbolModuleList=moduleMappingID(uniprotModuleList)names(symbolModuleList)=paste0("M",1:length(symbolModuleList))head(symbolModuleList)moonlightingCoefcient13moonlightingCoefficientmoonlightingCoefcientDescriptionCalculatethemoonlightingcoefcientforRNAs.
UsagemoonlightingCoefficient(rna2mod,simMat)Argumentsrna2modtwo-columnmatrixwitheachrowshowsthebinaryassociationbetweenRNAandprotein.
simMatadjacentmatrixindicatingthesemanticsimilarityamongproteinmodulesValuernaStatatableincludingncRNAsymble,numberoftargetmodules,andcorespondingmoonlightingcoefcient.
Author(s)LixinChengReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(rna2module)data(simMatBP.
Rel)mcOutput=moonlightingCoefficient(rna2module,simMat=simMatBP.
Rel)head(mcOutput)14pcdqModuleListpcdqModuleListpcdqModuleListDescriptionalistofmodulescollectedfromdatabasePCDq.
Usagedata("pcdqModuleList")FormatTheformatis:Listof412$pcdq_1:chr[1:39]"TPP1""A2M""GBA""GAPDH".
.
.
$pcdq_2:chr[1:41]"IKBKB""CHUK""MAP3K7""TUBB4A".
.
.
$pcdq_3:chr[1:24]"MED7""MED7""MED14""MED6".
.
.
$pcdq_4:chr[1:12]"BRCA1""MYC""ESR1""TP53".
.
.
$pcdq_5:chr[1:23]"PIK3R2""ABL1""EGFR""EGF".
.
.
$pcdq_6:chr[1:19]"FOS""FOS""JUN""FOSL1".
.
.
$pcdq_7:chr[1:14]"SF3B1""CD2BP2""U2AF2""YWHAG".
.
.
$pcdq_8:chr[1:10]"KAT7""VIM""SAT1""CHD3".
.
.
$pcdq_9:chr[1:6]"RTCA""HLA-B""IMPDH2""MCM3".
.
.
$pcdq_10:chr[1:18]"ZNHIT1""YEATS4""ACTL6A""MORF4L2".
.
.
$pcdq_11:chr[1:23]"PSMA7""EIF1B""EIF1B""EPB41".
.
.
$pcdq_12:chr[1:14]"PSMD11""SMAD7""SMAD9""SMAD6".
.
.
$pcdq_13:chr[1:13]"NCK1""SFPQ""SFPQ""PDIA3".
.
.
$pcdq_14:chr[1:17]"ORC5""ORC5""ORC4""CDK1".
.
.
$pcdq_15:chr[1:7]"APOA1""TTR""ALB""TF".
.
.
$pcdq_16:chr[1:7]"HGS""STAM2""DAZAP2""UBQLN4".
.
.
$pcdq_17:chr[1:6]"LDOC1""HNRNPC""FXR2""CCDC85B".
.
.
$pcdq_18:chr[1:7]"PLSCR1""SPRY2""GLRX3""EFEMP2".
.
.
$pcdq_19:chr[1:7]"BCL2""BID""BAX""BCL2L1".
.
.
$pcdq_20:chr[1:7]"GH1""RPL3""RPL3""EIF6".
.
.
$pcdq_21:chr[1:11]"CFLAR""FAS""FASLG""FASLG".
.
.
$pcdq_22:chr[1:10]"PIAS1""TP53""AR""SUMO1".
.
.
$pcdq_23:chr[1:12]"RB1""MYBL1""MYBL2""RBL2".
.
.
$pcdq_24:chr[1:8]"FYB1""FYN""HCK""SRC".
.
.
$pcdq_25:chr[1:12]"TNF""TNF""TNFRSF1A""TNFRSF1B".
.
.
$pcdq_26:chr[1:8]"FAM107A""FAM107A""KRT19""KRT15".
.
.
$pcdq_27:chr[1:10]"RBFOX2""CCNK""ATXN1""DAZAP2".
.
.
$pcdq_28:chr[1:13]"WDR5""RBX1""TLE2""CUL4A".
.
.
$pcdq_29:chr[1:24]"PSMD11""PSMD12""PSMD14""PSMD3".
.
.
$pcdq_30:chr[1:5]"SPTBN1""SP-TAN1""CEP63""EXOC1".
.
.
$pcdq_31:chr[1:20]"GEMIN2""SNRPB""SNRPB""GEMIN4".
.
.
$pcdq_32:chr[1:11]"ORC4""MCM3""MCM4""MCM5".
.
.
$pcdq_33:chr[1:16]"DXO""ZFP36""EXOSC10""EXOSC9".
.
.
$pcdq_34:chr[1:6]"CTNND1""CDH1""CTNNA1""CTNNB1".
.
.
$pcdq_35:chr[1:11]"RAD21""MYC""SMC1A""PDS5A".
.
.
$pcdq_36:chr[1:9]"CDC42""RAC1""YWHAH""PRKCZ".
.
.
$pcdq_37:chr[1:6]"RFXANK""HN-RNPM""C20orf24""NUP93".
.
.
$pcdq_38:chr[1:7]"MCC""VHL""ELOC""CUL2".
.
.
$pcdq_39:chr[1:14]"POLR2D""POLR2C""POLR2C""POLR2E".
.
.
$pcdq_40:chr[1:11]"BRCA1""NBN""CHEK2""H2AFX".
.
.
$pcdq_41:chr[1:8]"NUP214""NUP62""NUP153""RANBP2".
.
.
$pcdq_42:chr[1:8]"IFNAR1""TYK2""IL6ST""IL6ST".
.
.
$pcdq_43:chr[1:18]"ARID1A""ACTL6A""SMARCA2""SMARCA4".
.
.
$pcdq_44:chr[1:64]"PRMT5""RAD51B""HIST1H2BK""HIST1H2BJ".
.
.
$pcdq_45:chr[1:9]"HRAS""RAF1""BRAF""MAPK1".
.
.
$pcdq_46:chr[1:7]"TUBG1""AKT1""AKT2""GSK3A".
.
.
$pcdq_47:chr[1:8]"THRB""VDR""RXRA""PRMT2".
.
.
$pcdq_48:chr[1:8]"MEN1""RPA2""RPA1""RPA3".
.
.
$pcdq_49:chr[1:8]"APAF1""CASP1""CASP3""CASP9".
.
.
$pcdq_50:chr[1:9]"BACH1"pcdqModuleList15"CREB3""CREB1""CREB1".
.
.
$pcdq_51:chr[1:7]"TXNDC9""PTN""CSTF2""BAG6".
.
.
$pcdq_52:chr[1:20]"SAP18""HDAC3""NCOR1""SAP30".
.
.
$pcdq_53:chr[1:5]"GFI1B""ACTN1""TRIM27""MAGEA11".
.
.
$pcdq_54:chr[1:13]"ARPC1B""ARPC2""ARPC2""ARPC3".
.
.
$pcdq_55:chr[1:7]"KPNA3""ARRB2""ARRB2""CSNK2A1".
.
.
$pcdq_56:chr[1:7]"TP53""HTT""NAP1L1""CTBP1".
.
.
$pcdq_57:chr[1:9]"EIF4G3""EIF4E""EIF4E""EIF4A2".
.
.
$pcdq_58:chr[1:10]"SNRNP200""MYC""HNRNPC""TOP1".
.
.
$pcdq_59:chr[1:9]"FANCG""FANCG""FANCA""FANCC".
.
.
$pcdq_60:chr[1:7]"DVL2""ZNF250""FXR2""AP2M1".
.
.
$pcdq_61:chr[1:6]"SNAPIN""BLOC1S1""BLOC1S4""BLOC1S2".
.
.
$pcdq_62:chr[1:8]"DDX6""ZFP36""DCP1A""EDC4".
.
.
$pcdq_63:chr[1:7]"GDF9""ZNF24""APLP1""EEF1A1".
.
.
$pcdq_64:chr[1:13]"IRAK2""IL1R1""IRAK1""TIRAP".
.
.
$pcdq_65:chr[1:10]"COPS2""GPS1""COPS4""COPS3".
.
.
$pcdq_66:chr[1:6]"SMC2""NCAPH""NCAPD2""NEK6".
.
.
$pcdq_67:chr[1:7]"WWP2""EWSR1""HERPUD1""HER-PUD1".
.
.
$pcdq_68:chr[1:14]"PPP2R1A""PPP2R1B""PPP2CB""PPP2R2A".
.
.
$pcdq_69:chr[1:5]"GAS7""NCKAP1""ABI1""CYFIP2".
.
.
$pcdq_70:chr[1:9]"CBL""CBLB""PDCD6IP""SH3KBP1".
.
.
$pcdq_71:chr[1:9]"CDK4""CCND1""CCND1""CCND3".
.
.
$pcdq_72:chr[1:11]"ACTL6A""TCF3""RUVBL2""INO80C".
.
.
$pcdq_73:chr[1:10]"LSM1""LSM2""LSM8""SNRPE".
.
.
$pcdq_74:chr[1:25]"SF3B1""SNRPB""SNRPB""U2AF2".
.
.
$pcdq_75:chr[1:5]"BCR""CFTR""COPB1""PDZK1".
.
.
$pcdq_76:chr[1:7]"TMSB4X""LIMS1""ERH""ILK".
.
.
$pcdq_77:chr[1:6]"GNAI2""FLNA""CANX""MTNR1A".
.
.
$pcdq_78:chr[1:6]"TPD52L2""TPD52L2""TPD52""TPD52L1".
.
.
$pcdq_79:chr[1:9]"KRT6A""KRT6C""KRT18""KRT8".
.
.
$pcdq_80:chr[1:7]"ERCC2""ERCC3""ERCC5""GTF2H1".
.
.
$pcdq_81:chr[1:5]"DNMT1""DNMT1""SUZ12""EZH2".
.
.
$pcdq_82:chr[1:9]"TGFB1""TGFB3""ENG""ENG".
.
.
$pcdq_83:chr[1:6]"CRYAA""CRYAA2""CRYAB""HSPB1".
.
.
$pcdq_84:chr[1:6]"MAFG""MAFK""JUN""NFE2L2".
.
.
$pcdq_85:chr[1:20]"ITGB3""ITGB1""ITGAV""ITGA5".
.
.
$pcdq_86:chr[1:8]"PARP1""XRCC6""XRCC5""XRCC1".
.
.
$pcdq_87:chr[1:7]"CDC37""CCT7""TANK""TANK".
.
.
$pcdq_88:chr[1:9]"CBX4""E2F6""BMI1""COMMD3-BMI1".
.
.
$pcdq_89:chr[1:8]"CDK11B""CDK11A""YWHAQ""YWHAB".
.
.
$pcdq_90:chr[1:6]"PEX14""ABCD3""ABCD1""PEX19".
.
.
$pcdq_91:chr[1:10]"POP7""RPP14""POP4""RPP38".
.
.
$pcdq_92:chr[1:7]"UBE2M""NEDD8""CAV1""CAV1".
.
.
$pcdq_93:chr[1:6]"F2""VTN""ITGB3""SERPINE1".
.
.
$pcdq_94:chr[1:5]"CD4""NR3C1""LCK""HSP90AA1".
.
.
$pcdq_95:chr[1:10]"NDC80""ZWINT""CBX5""CBX3".
.
.
$pcdq_96:chr[1:6]"ASB1""ASB6""ASB12""CUL5".
.
.
$pcdq_97:chr[1:7]"MYC""PCNA""RFC4""RFC2".
.
.
$pcdq_98:chr[1:7]"MEN1""WDR5""KMT2A""RBBP5".
.
.
$pcdq_99:chr[1:6]"TOM1L1""PTBP1""HNRNPH1""CRMP1".
.
.
[listoutputtruncated]ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(pcdqModuleList)head(pcdqModuleList)16predModuleListppippiDescriptiontwo-colomnmatrixwitheachrowshowsinteractionbetweenapairofproteins.
Usagedata("ppi")FormatTheformatis:chr[1:210410,1:2]"KANSL1L""SLC12A8""RBM47""RBM47""RBM47""RBM47""RBM47""RBM47""TTC26""TTC26""TTC26".
.
.
-attr(*,"dimnames")=Listof2.
.
$:NULL.
.
$:chr[1:2]"V1""V2"ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(ppi)head(ppi)predModuleListpredictedmodulesDescriptionalistofproteinmoudlesidentiedusingClusterONE.
Usagedata("predModuleList")FormatTheformatis:Listof837$pred_1:chr[1:216]"FBXO4""FBXL3""FBXW2""ISY1".
.
.
$pred_2:chr[1:169]"TNRC6A""SH3RF2""GPX8""SAP25".
.
.
$pred_3:chr[1:17]"NXT1""NUDT5""IKZF2""FBXO5".
.
.
$pred_4:chr[1:228]"PDK3""MAPRE1""TSC22D1""ELF2".
.
.
$pred_5:chr[1:51]"POLR2F""PSME3""MAGOH""VBP1".
.
.
$pred_6:chr[1:17]"FAU""RPL23""UBE2D2""RPS15".
.
.
$pred_7:chr[1:559]"RPL36AL""L3MBTL2""MUL1""RNF166".
.
.
$pred_8:chr[1:22]"NUBP1""RAD23A""RAD23B""EIF5".
.
.
$pred_9:chr[1:60]"MAK16""NIFK""GTPBP4""WDR33".
.
.
$pred_10:chr[1:296]"DYRK3""NXNL2"predModuleList17"ANKS6""NAIF1".
.
.
$pred_11:chr[1:10]"EHBP1""GPKOW""SNAPC3""CLP1".
.
.
$pred_12:chr[1:144]"ZNF165""PAPOLA""BRCA2""PSMD7".
.
.
$pred_13:chr[1:46]"DAAM2""PRPF39""RPAIN""KTN1".
.
.
$pred_14:chr[1:863]"FANCL""RBFOX1""BABAM1""IRAK4".
.
.
$pred_15:chr[1:80]"TRA2B""UBE2B""PPP2R2A""SNRPN".
.
.
$pred_16:chr[1:77]"LMX1A""TOPBP1""ZNF516""DHX38".
.
.
$pred_17:chr[1:63]"ZNF587B""NACA""PRPF4B""HDAC1".
.
.
$pred_18:chr[1:253]"ELP5""LMX1A""SH3RF2""GPX8".
.
.
$pred_19:chr[1:45]"RELB""GALK2""AMPD2""CTBS".
.
.
$pred_20:chr[1:6]"SMG9""NAPB""UPF3A""WNT8A".
.
.
$pred_21:chr[1:69]"ZNF45""ZNF33A""ZNF33B""PPARA".
.
.
$pred_22:chr[1:430]"NOL8""DHX30""EIF3M""MICAL3".
.
.
$pred_23:chr[1:186]"NPAT""EIF2B1""EIF4A2""TCEB3".
.
.
$pred_24:chr[1:315]"MACROD1""PAAF1""RIOK1""LARP6".
.
.
$pred_25:chr[1:6]"DDX18""SDAD1""NLE1""RBM28".
.
.
$pred_26:chr[1:56]"HNRNPH3""HNRNPH1""STIP1""ARRB2".
.
.
$pred_27:chr[1:43]"WDR1""TDO2""POLD2""ALDH9A1".
.
.
$pred_28:chr[1:14]"SLC25A16""GLB1""PPP3CB""PFKL".
.
.
$pred_29:chr[1:32]"EXOSC4""ISYNA1""NOX4""DCP1A".
.
.
$pred_30:chr[1:296]"NACA""PRPF4B""HDAC1""TDG".
.
.
$pred_31:chr[1:199]"TSC22D1""NAB2""TAB1""MAPK11".
.
.
$pred_32:chr[1:33]"LTA4H""HSPA1A""HSPA1B""U2AF1L5".
.
.
$pred_33:chr[1:6]"RBMX""IFNGR2""EIF4A3""IGHMBP2".
.
.
$pred_34:chr[1:37]"CCT8""CCT4""ANXA11""PAPOLA".
.
.
$pred_35:chr[1:12]"GLTSCR1L""ZMAT3""CWC22""XAB2".
.
.
$pred_36:chr[1:183]"TMEM62""FAAP100""CPSF1""KIF1A".
.
.
$pred_37:chr[1:99]"HBG2""PSPH""RBM6""RPP30".
.
.
$pred_38:chr[1:214]"SSBP2""ERH""RPL19""SRSF3".
.
.
$pred_39:chr[1:60]"USF1""ME2""GLDC""WARS".
.
.
$pred_40:chr[1:17]"NAT10""DDX47""ESF1""HEATR1".
.
.
$pred_41:chr[1:134]"RNH1""RARG""ANXA8""ENO3".
.
.
$pred_42:chr[1:13]"ZNF251""ZNF2""ZNF426""ZNF416".
.
.
$pred_43:chr[1:25]"METTL25""DDX51""KRI1""SRFBP1".
.
.
$pred_44:chr[1:21]"EIF4A1""RPS20""PRPS1""SHFM1".
.
.
$pred_45:chr[1:23]"SMC4""MAN2C1""SACM1L""OLA1".
.
.
$pred_46:chr[1:55]"BRPF1""EIF6""SIK1""WDR4".
.
.
$pred_47:chr[1:62]"PDHA2""BLVRB""PRDX5""PEBP1".
.
.
$pred_48:chr[1:24]"CNGB1""GAPVD1""HERPUD1""LAPTM4A".
.
.
$pred_49:chr[1:31]"TRIM2""FAM117A""INO80B""CCSER2".
.
.
$pred_50:chr[1:7]"RBM22""CWC25""CWC15""CPSF2".
.
.
$pred_51:chr[1:6]"ZNF160""VTA1""ZNF331""CELSR1".
.
.
$pred_52:chr[1:8]"ZNF324B""ZNF470""ZNF425""ZNF529".
.
.
$pred_53:chr[1:9]"ZNF225""ZNF221""ZNF510""ZNF256".
.
.
$pred_54:chr[1:65]"ISYNA1""NOX4""RIC8A""RRAGD".
.
.
$pred_55:chr[1:37]"SSR1""NOP2""NSF""MAP2K3".
.
.
$pred_56:chr[1:29]"PSMB10""RBM34""IL12RB1""CDKN2B".
.
.
$pred_57:chr[1:58]"POM121C""PODXL""KIF3C""NRP1".
.
.
$pred_58:chr[1:26]"WDR61""INTS2""NELFA""TAF1D".
.
.
$pred_59:chr[1:20]"GNL1""LTBR""GPX4""HSD17B3".
.
.
$pred_60:chr[1:14]"RFC4""RFC2""RFC1""GTF2F1".
.
.
$pred_61:chr[1:14]"U2AF1L4""PPIL4""CCDC12""LEO1".
.
.
$pred_62:chr[1:8]"MRPS28""MRPS7""MRPS2""COQ4".
.
.
$pred_63:chr[1:25]"SUPT20H""ARMC2""NCOA7""SLC30A5".
.
.
$pred_64:chr[1:26]"MAPK3""MARK3""RPA1""APEX1".
.
.
$pred_65:chr[1:27]"MAST3""RASAL1""SNAPIN""PAK4".
.
.
$pred_66:chr[1:7]"STBD1""FEN1""MLH1""IL15".
.
.
$pred_67:chr[1:34]"STMN1""YBX3""ZFX""RHOQ".
.
.
$pred_68:chr[1:23]"RINT1""BZW1""NOXA1""LAPTM4B".
.
.
$pred_69:chr[1:61]"RAMP1""BCAS2""DNAJC8""SMNDC1".
.
.
$pred_70:chr[1:121]"MLXIPL""ISYNA1""NOX4""DCP1A".
.
.
$pred_71:chr[1:49]"MAN1A1""SPR""CBS""GCK".
.
.
$pred_72:chr[1:14]"ZNF721""ZFP90""ZNF473""ZNF480".
.
.
$pred_73:chr[1:16]"ZNF433""ZNF283""CNTD1""ZNF614".
.
.
$pred_74:chr[1:69]"RBM25""EIF2B2""HINT1""STK19".
.
.
$pred_75:chr[1:24]"SULT1C4""EPRS""HSP90AA1""GALT".
.
.
$pred_76:chr[1:8]"AHI1""HUS1B""ALMS1""CCHCR1".
.
.
$pred_77:chr[1:75]"ALB""MAP4K3""TBC1D2""NUF2".
.
.
$pred_78:chr[1:37]"C11orf49""TMEM231""C14orf93""ZMAT3".
.
.
$pred_7918rna2mod:chr[1:21]"CFB""LHB""PRL""GCG".
.
.
$pred_80:chr[1:19]"GIPC3""GIPC2""BTBD6""LRR1".
.
.
$pred_81:chr[1:12]"ZNF667""ZNF484""ZNF569""ZNF684".
.
.
$pred_82:chr[1:23]"MRPS5""MRPS36""MRPS11""MRPS15".
.
.
$pred_83:chr[1:14]"DDX47""ESF1""HEATR1""PIF1".
.
.
$pred_84:chr[1:12]"C14orf93""FBRS""GPBP1L1""ZNF532".
.
.
$pred_85:chr[1:11]"ZNF605""ZNF554""ZNF429""ZNF677".
.
.
$pred_86:chr[1:9]"ZNF251""ZNF2""ZNF426""EFCAB11".
.
.
$pred_87:chr[1:12]"RMI1""CLSPN""FANCE""UBE2T".
.
.
$pred_88:chr[1:40]"LGALSL""PARP14""PLEKHN1""ZC2HC1C".
.
.
$pred_89:chr[1:48]"TSPAN2""TSPAN3""SNAP91""RAMP2".
.
.
$pred_90:chr[1:8]"NUP37""NUP35""CASC5""FBXO30".
.
.
$pred_91:chr[1:12]"APOL2""LBHD1""ACBD6""ZFP14".
.
.
$pred_92:chr[1:29]"SMG9""NAPB""UPF3A""WNT8A".
.
.
$pred_93:chr[1:33]"EIF4E""ENO1""PYGL""GPI".
.
.
$pred_94:chr[1:11]"XDH""RFX3""PIP4K2A""SOX2".
.
.
$pred_95:chr[1:26]"COX20""RBM48""CELF3""SZT2".
.
.
$pred_96:chr[1:14]"ZNF613""ZNF790""ZNF562""ZNF782".
.
.
$pred_97:chr[1:8]"SNAPC4""CPSF3L""RPRD2""TAF3".
.
.
$pred_98:chr[1:24]"ZNF821""DAB1""PLCG2""KLRC2".
.
.
$pred_99:chr[1:15]"RANGAP1""NUP153""RANBP2""GSK3A".
.
.
[listoutputtruncated]ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(predModuleList)head(predModuleList)rna2modrna2modDescriptionConstructa0-1binarymatrixtorepresenttheassociationbetweenRNAsandproteinmodules.
Usagerna2mod(rna2prot,modlist,pCutoff=0.
01,bgProtNum)Argumentsrna2prottwo-columnmatrixwitheachrowshowsthebinaryassociationbetweenRNAandprotein.
modlistalistofproteinmodules.
pCutoffthresholdoftheP-valueforenrichmentanalysis.
bgProtNumthenumberofbackgroundproteins.
Valuerna2moduleNxM0-1binarymatrixofNRNAandMmodule.
moduleListalistofmodulestargetedbyatleastoneRNA.
rna2module19Author(s)LixinChengReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(rna2protein)data(combinedModuleList)data(uniGene)uniGeneNum=length(uniGene)result=rna2mod(rna2protein[1:100,],combinedModuleList,pCutoff=0.
01,uniGeneNum)rna2module=result[[1]]moduleList=result[[2]]rna2moduleRNA-moduleassociationmatrixDescriptionA0-1binarymatrixrepresentingtheassociationbetweenRNAsandmodulesUsagedata("rna2module")FormatTheformatis:num[1:776,1:175]0000000000.
.
.
-attr(*,"dimnames")=Listof2.
.
$:chr[1:776]"CRNDE""NEAT1""ZFAS1""GAS5"chr[1:175]"1""2""3""4".
.
.
ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(rna2module)head(rna2module)20SDIrna2proteinrna2proteinDescriptiontwo-columnmatrixwitheachrowshowsthebinaryassociationbetweenRNAandprotein.
Usagedata("rna2protein")FormatTheformatis:chr[1:12008,1:2]"LINC00271""MALAT1""CRNDE""PINK1-AS""TUG1""MALAT1""CRNDE""C11orf95""ANCR""LOC647323".
.
.
-attr(*,"dimnames")=Listof2.
.
$:NULL.
.
$:chr[1:2]"Interactor1""Interactor2"ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(rna2protein)head(rna2protein)SDIShannondiversityindexDescriptionCalculatetheSimpson'sdiversityindex(SDI).
UsageSDI(x)Argumentsxweightsoflatentfeatures.
Valuezmoonlightingcoefcient,ornormalizedShannon'sdiversityindex.
simMatBP.
Jiang21Author(s)LixinChengReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesa=sample(1:10,4)b=a/sum(a)SDI(b)simMatBP.
Jiangasemanticsimilarity(Jiang)matrixDescriptionAnadjacentmatrixofthesemanticsimilarity(Jiang)amongdiffernetproteinmodules.
Usagedata("simMatBP.
Jiang")FormatTheformatis:num[1:175,1:175]10.
8570.
9350.
8680.
5470.
780.
7130.
7750.
7790.
807.
.
.
-attr(*,"dimnames")=Listof2.
.
$:chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4"chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4".
.
.
ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(simMatBP.
Jiang)dim(simMatBP.
Jiang)22simMatBP.
RelsimMatBP.
Linasemanticsimilarity(Lin)matrixDescriptionAnadjacentmatrixofthesemanticsimilarity(Lin)amongdiffernetproteinmodules.
Usagedata("simMatBP.
Lin")FormatTheformatis:num[1:175,1:175]10.
8770.
9450.
8860.
5610.
8080.
7330.
7930.
8050.
825.
.
.
-attr(*,"dimnames")=Listof2.
.
$:chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4"chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4".
.
.
ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(simMatBP.
Lin)head(simMatBP.
Lin)simMatBP.
Relasemanticsimilarity(Rel)matrixDescriptionAnadjacentmatrixofthesemanticsimilarity(Rel)amongdiffernetproteinmodules.
Usagedata("simMatBP.
Rel")FormatTheformatis:num[1:175,1:175]0.
9940.
870.
9390.
8790.
5340.
7980.
720.
7840.
7950.
815.
.
.
-attr(*,"dimnames")=Listof2.
.
$:chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4"chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4".
.
.
ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
simMatBP.
Resnik23Examplesdata(simMatBP.
Rel)head(simMatBP.
Rel)simMatBP.
Resnikasemanticsimilarity(Resnik)matrixDescriptionAnadjacentmatrixofthesemanticsimilarity(Resnik)amongdiffernetproteinmodules.
Usagedata("simMatBP.
Resnik")FormatTheformatis:num[1:175,1:175]0.
5860.
4980.
5420.
5050.
2960.
4550.
4010.
4420.
4510.
46.
.
.
-attr(*,"dimnames")=Listof2.
.
$:chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4"chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4".
.
.
ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(simMatBP.
Resnik)head(simMatBP.
Resnik)simMatBP.
Wangasemanticsimilarity(Wang)matrix~DescriptionAnadjacentmatrixofthesemanticsimilarity(Wang)amongdiffernetproteinmodules.
Usagedata("simMatBP.
Wang")FormatTheformatis:num[1:175,1:175]10.
8270.
9190.
8450.
5270.
7610.
6590.
7250.
7420.
77.
.
.
-attr(*,"dimnames")=Listof2.
.
$:chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4"chr[1:175]"mapped_1""mapped_2""mapped_3""mapped_4".
.
.
24uniGeneReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(simMatBP.
Wang)head(simMatBP.
Wang)uniGeneBackgroundgenesymbolsDescriptionAllthegenesymbolsusedintheentireprotieninteractionnetwork.
Usagedata("uniGene")FormatTheformatis:chr[1:10111]"A1CF""AAAS""AADAT""AAGAB""AAK1""AAMDC""AAMP""AAR2""AARS""AARS2""AARSD1""AASDHPPT".
.
.
ReferencesMoonFinder:aframeworkfortheidenticationofmoonlightingnon-codingRNAs.
Examplesdata(uniGene)head(uniGene)IndexTopicdatasetscombinedModuleList,3corumModuleList,5moduleList,10pcdqModuleList,14ppi,16predModuleList,16rna2module,19rna2protein,20simMatBP.
Jiang,21simMatBP.
Lin,22simMatBP.
Rel,22simMatBP.
Resnik,23simMatBP.
Wang,23uniGene,24TopicpackageMoonFinder-package,2combinedModuleList,3corumModuleList,5moduleCombiding,7moduleFunctionComparing,8moduleHeatmap,9moduleList,10moduleMappingID,12MoonFinder(MoonFinder-package),2MoonFinder-package,2moonlightingCoefficient,13pcdqModuleList,14ppi,16predModuleList,16rna2mod,18rna2module,19rna2protein,20SDI,20simMatBP.
Jiang,21simMatBP.
Lin,22simMatBP.
Rel,22simMatBP.
Resnik,23simMatBP.
Wang,23uniGene,2425
- CHEK2ww.00271.com相关文档
- plateletww.00271.com
- movementsww.00271.com
- openww.00271.com
- Currentww.00271.com
- restrictingww.00271.com
- Mermaidww.00271.com
sharktech:洛杉矶/丹佛/荷兰高防服务器;1G独享$70/10G共享$240/10G独享$800
sharktech怎么样?sharktech (鲨鱼机房)是一家成立于 2003 年的知名美国老牌主机商,又称鲨鱼机房或者SK 机房,一直主打高防系列产品,提供独立服务器租用业务和 VPS 主机,自营机房在美国洛杉矶、丹佛、芝加哥和荷兰阿姆斯特丹,所有产品均提供 DDoS 防护。不知道大家是否注意到sharktech的所有服务器的带宽价格全部跳楼跳水,降幅简直不忍直视了,还没有见过这么便宜的独立服...
BuyVM迈阿密KVM上线,AMD Ryzen 3900X+NVMe硬盘$2/月起
BuyVM在昨天宣布上线了第四个数据中心产品:迈阿密,基于KVM架构的VPS主机,采用AMD Ryzen 3900X CPU,DDR4内存,NVMe硬盘,1Gbps带宽,不限制流量方式,最低$2/月起,支持Linux或者Windows操作系统。这是一家成立于2010年的国外主机商,提供基于KVM架构的VPS产品,数据中心除了新上的迈阿密外还包括美国拉斯维加斯、新泽西和卢森堡等,主机均为1Gbps带...
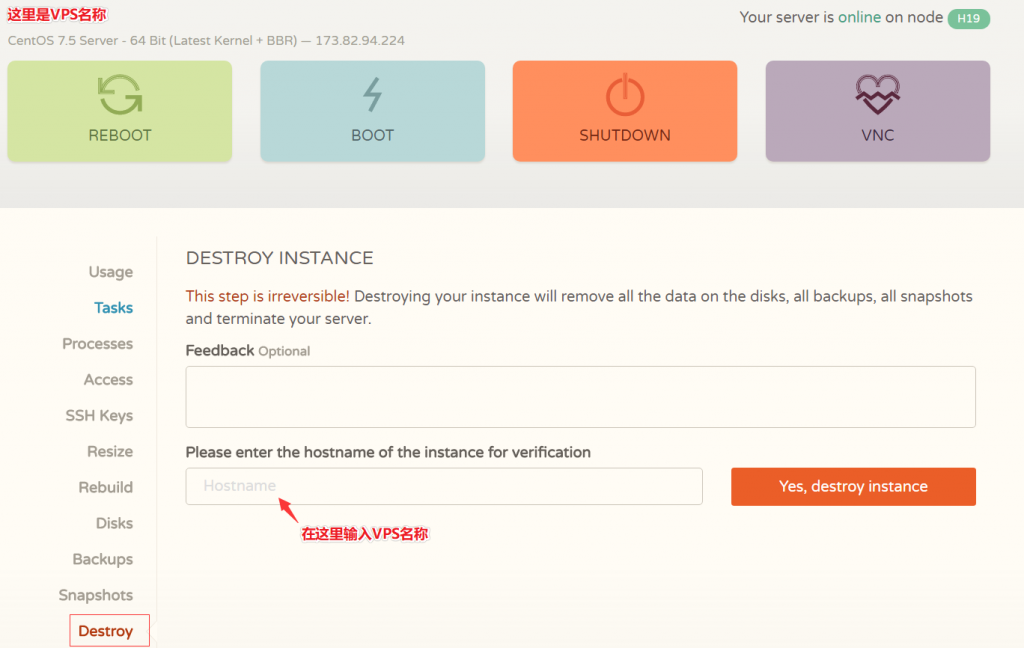
CloudCone(20美元/年)大硬盘VPS云服务器,KVM虚拟架构,1核心1G内存1Gbps带宽
近日CloudCone商家对旗下的大硬盘VPS云服务器进行了少量库存补货,也是悄悄推送了一批便宜VPS云服务器产品,此前较受欢迎的特价20美元/年、1核心1G内存1Gbps带宽的VPS云服务器也有少量库存,有需要美国便宜大硬盘VPS云服务器的朋友可以关注一下。CloudCone怎么样?CloudCone服务器好不好?CloudCone值不值得购买?CloudCone是一家成立于2017年的美国服务...
ww.00271.com为你推荐
-
编程小学生惊库克小学生学编程好吗?美国互联网瘫痪美国网络大瘫痪到底是怎么发生的摩拜超15分钟加钱首次 微信扫 摩拜单车 需要 付压金吗今日油条天天吃油条,身体会怎么样刘祚天DJ这个职业怎么样?同ip网站同IP网站9个越来越多,为什么?百度指数词百度指数是指,词不管通过什么样的搜索引擎进行搜索,都会被算成百度指数吗?partnersonline我家Internet Explorer为什么开不起来ww.66bobo.com谁知道11qqq com被换成哪个网站本冈一郎本冈一郎有副作用吗?主要有什么呢?