samnajie
najie 时间:2021-02-28 阅读:()
ArchivesofVirology(2018)163:725–730https://doi.
org/10.
1007/s00705-017-3645-1BRIEFREPORTAcomparisonoftheMeltProHPVTestwiththeCobasHPVTestfordetectingandgenotyping14highriskhumanpapillomavirustypesZhitengTang1·YeXu2·NajieSong3·DongqingZou3·YiqunLiao4·QinggeLi2·ChaoPan1Received:11May2017/Accepted:3October2017/Publishedonline:5December2017Springer-VerlagGmbHAustria,partofSpringerNature2017AbstractTheclinicalperformanceofthenewlydevelopedMeltProHPVTest,basedonmulticolormeltingcurveanalysis,wasevaluatedandcomparedwiththecommerciallyavailableCobasHPVTestfordetectionofHPVandgenotypingofHPV-16andHPV-18.
Atotalof1647cervicalsampleswereanalyzedwithbothtests.
Theagreementvalueswere96.
2%forHPVdetection,99.
6%forHPV-16identification,and99.
7%forHPV-18identification.
AllgenotypingresultsfromMeltProHPVTestshowedthatHPV-52,HPV-58,andHPV-16werethemostcommontypesinthisstudy.
Intra-laboratoryreproducibilitystudiesshowed97.
8%agreementwhileinter-laboratoryreproducibilitystudiesshowed96.
9%agreementfortheMeltProHPVTest.
TheMeltProHPVTestandCobasHPVTestarehighlycorrelativeandareusefulformonitoringHPVinfection.
High-riskhumanpapillomavirus(HPV)infectionisahighriskfactorforthedevelopmentofcervicalcancer,whichisthesecondmostcommonmalignanttumorinwomenworld-wide[1].
High-riskHPVdiagnosescombinedwithliquid-basedcytologyanalysisisconsideredtobethemosteffectivemethodforearlycervicalcancerscreening[2].
Thefirstedi-tionofthe"Humanpapillomaviruslaboratorymanual"waspublishedin2009bytheWorldHealthOrganization(WHO)toprovideguidanceforhigh-riskHPVdiagnoses[3].
HPVvaccinationiscurrentlythesafestapproachforpreventingthedevelopmentofcervicalcancer[4].
ThedistributionofHPVtypeandtheindividualriskofeachHPVtypearetwoimportantfactorsthatneededtobeconsideredfordevelopingtheHPVvaccineagainstparticularHPVgenotypeswithinacountry[5].
Convenientandaccuratetechniquesforhigh-riskHPVdetectionandgenotypingareurgentlyneededforHPVclinicaldiagnosesandepidemiologicalstudies.
ThehybridizationcapturedetectionforHPVgeneticDNAoritsPCRampliconisatraditionalapproachforHPVgeno-typing[6–9].
However,itrequiresmanyoperationalsteps,andPCRcontaminationisacommonproblem.
Avarietyofmethodsbasedonreal-timePCRhavebeendevelopedintheHandlingEditor:ZhongjieShi.
ZhitengTangandYeXucontributedequallytothiswork.
ElectronicsupplementarymaterialTheonlineversionofthisarticle(https://doi.
org/10.
1007/s00705-017-3645-1)containssupplementarymaterial,whichisavailabletoauthorizedusers.
*YiqunLiaoyqliao@xmu.
edu.
cn*QinggeLiqgli@xmu.
edu.
cn*ChaoPanpanchao123a@126.
com1ZhongshanHospital,XiamenUniversity,Xiamen,Fujian,China2TheStateKeyLaboratoryofCellularStressBiology,StateKeyLaboratoryofMolecularVaccinologyandMolecularDiagnostics,EngineeringResearchCenterofMolecularDiagnosticsoftheMinistryofEducation,SchoolofLifeSciences,XiamenUniversity,Xiamen,Fujian,China3ZeesanBiotechnologyCompany,Xiamen,Fujian,China4TheStateKeyLaboratoryofMolecularVaccinologyandMolecularDiagnostics,StateKeyLaboratoryofCellularStressBiology,EngineeringResearchCenterofMolecularDiagnosticsoftheMinistryofEducation,SchoolofPublicHealth,XiamenUniversity,Xiamen,Fujian,China726Z.
Tangetal.
pastdecade[10–13].
Comparedwithtraditionalhybridizationcapturedetection,thereal-timePCRplatformpossessestheadvantagesofconvenience,highthroughput,lowtimeandcost,andlowriskoffalse-positiveresultsduetocross-con-taminationofPCR.
Commercialreal-timePCRkitsarenowwidelyadoptedinclinicalHPVdiagnosticandresearchstud-ies.
TheCobasHPVTestisatypicalsystemthathasbeenapprovedbytheU.
S.
FoodandDrugAdministration(FDA).
InapreviousstudywereportedanovelHPVgenotyp-ingmethodbasedonreal-timePCRandmeltcurveanalysis[14].
TheMeltProHPVTestwasdevelopedwiththesamemethod,andcandetectandgenotypethe14mostcommonhigh-riskHPVtypesinasingle-tubereaction.
Inthisstudy,weperformedacomparisonoftheMeltProHPVTestandtheCobasHPVTestforthedetectionandgenotypingof14high-riskHPVtypesinsoutheastChina.
TheprocessesinvolvedinthiscomparisonstudyareshowninFig.
S1.
More-over,reproducibilityisimportantforanynewlydevelopedmethod.
AprotocolforclinicalvalidationofHPVassays,called"VALGENT",wasdevelopedandsuccessfullyappliedforthecomparisonofmanycommercialHPVtests[15–18].
Followingthisguideline,intra-laboratoryandinter-laboratoryreproducibilityexperimentswereperformedinthisstudy.
ForcomparisonoftheMeltProHPVTestandtheCobasHPVTest,atotalof1647residualcervixcellsam-pleswerecollectedfromindividualwomenlivinginthesoutheastofChina.
Pregnantwomenwereexcludedfromthisstudy.
AllsampleswerecollectedatZhongshanHos-pital,XiamenUniversity(Xiamen,Fujian,China)withaThinPrepliquid-basedcytologysystem,in2015,andstoredat-20°Cfor2weeksbeforeanalysis.
Theageofthepatientsisfrom19to65yearsoldwithameanageof32years-oldandamedianof31years-old.
AlloftheexperimentsinthiscomparisonstudywereperformedatZhongshanHospital.
Fortheintra-laboratoryandinter-laboratoryreproducibilitystudyoftheMeltProHPVTest,atotalof540residualcervixcellsampleswerecollectedatZhongshanhospitalbythesameprotocolin2017.
Theintra-laboratoryreproduc-ibilitystudywasperformedatZhongshanHospital,whiletheinter-laboratoryreproducibilitiesstudywasperformedatZhongshanHospitalandtheEngineeringResearchCenterofMolecularDiagnosticsoftheMinistryofEducation,Xia-menUniversity.
ThestudyprotocolwasapprovedbyTheResearchEthicsCommitteeofXiamenUniversity.
Forthecomparisonstudy,1647sampleswereassayedinadouble-blindedfashionusingtheMeltProHPVTest(ZeesanBiotechnologyCo.
,China)andtheCobasHPVTest(RocheDiagnosticsCo.
,Switzerland)onthesameday.
DNAextrac-tionwasautomaticallyperformed,separately,accordingtotheprotocolsofthesetwosystems.
TheMeltProHPVTestdetected14high-riskHPVtypes(HPV-16,18,31,33,35,39,45,51,52,56,58,59,66,and68)andprovidedspecificgeno-typingforall14HPVtypesbasedonmeltcurveanalysis.
Theglyceraldehyde-3-phosphatedehydrogenase(GAPDH)genewasusedasaninternalcontrolintheMeltProHPVTest.
ThePCRreactionanddataprocessingstepsoftheMeltProHPVTestwereperformedintheSLAN96real-timePCRsystem.
TheCobasHPVTestdetectedthesame14high-riskHPVtypesbutonlyprovidedspecificgenotypingforHPV-16andHPV-18,basedonreal-timePCR.
Theβ-globulingenewasusedasinternalcontrolintheCobasHPVTest.
ThePCRreactionanddataprocessingstepsoftheCobasHPVTestwereperformedinaCobas4800real-timePCRsystem.
TheexperimentalconditionsforTheMeltProHPVTestandCobasHPVTestfollowedtheguidelinesoftheirassociatedprotocols.
Duringeachrun,forbothassays,apositiveandnegativecontrolwasincludedtoensureproperPCRreactionsandthattherewasnocarry-overcontamination.
TheHPVgenotypeforeachsamplewasassayedusingindependent,automated-readoutsoftwaresuppliedwiththeMeltProHPVsystemorCobasHPVsystems.
Thediag-nosticresultsfromtheMeltProHPVTestandtheCobasHPVTestwerecomparedtoevaluateagreement.
Foralldis-crepantsamples,theoriginaldatawereprocessedmanually.
ForcaseswithdiscrepantresultsofHPV-16andHPV-18,confirmatorytestingwasperformedusinganHPVtype-specificreal-timePCRassaydiagnostickitdesignedonlyforHPV-16andHPV-18(KehuaLtd.
,Shanghai,China).
Fortheintra-laboratoryreproducibilitystudy,540sam-pleswereassayedbytheMeltProHPVTesttwiceonthesamedayatZhongshanhospital,andtheresultsofthefirstandsecondassaywerecompared.
Fortheinter-laboratoryreproducibilitystudy,thesame540sampleswereassayedoneweeklaterattheEngineeringResearchCenterofMolecularDiagnosticsoftheMinistryofEducation,andtheresultswerecomparedwiththefirstassay'sresultsfromtheintra-laboratoryreproducibilitystudy.
StatisticalanalysisforthecomparisondatawascarriedoutusingtheSPSSstatisticalsoftware(version13.
0,SPSSInc.
,Chicago,IL).
Epidemiologicalprevalence,forthe14high-riskHPVtypes,wasonlycalculatedfromresultsobtainedwiththeMeltProHPVTest,whichwascapableoffullgenotyping.
Wecomparedtheresultsobtainedfrom1647samplesusingtheMeltProHPVTestandtheCobasHPVTestforthedetectionof14high-riskHPVtypeswithoutgenotyping(Table1).
Theoverallagreementbetweenthesetestswas96.
2%(1584/1647),andthekappavaluewas0.
881(95%CI,0.
851-0.
911).
SincetheMeltProHPVTestandtheCobasHPVTestbothprovideHPV-16andHPV-18geno-typing,wealsoevaluatedthegenotypingresultsforHPV-16andHPV-18,withinthecomparisonshowninTable1.
TheagreementforthegenotypingresultsforHPV-16was99.
6%(1641/1647),andthekappavaluewas0.
946(95%CI,0.
902-0.
990).
TheagreementforthegenotypingresultsforHPV-18was99.
7%(1642/1647),andthekappavaluewas7270.
904(95%CI,0.
818-0.
990).
TheresultsfortheHPVdetec-tion,HPV-16genotypingandHPV-18genotypingusingtheMeltProHPVTestandtheCobasHPVTestwerenotsignificantlydifferent(McNemar'sTest,Pvalue=0.
23forHPVdetection,Pvalue=0.
69forHPV-16genotyping,Pvalue=1.
00forHPV-18genotyping).
Forthe63discrepantsamples,weprocessedtheorigi-naldatamanually.
ThequantitativePCRcycle(Cq)valuesforthe22samplesdiagnosedaspositivebytheCobasHPVTestandnegativebytheMeltProHPVTestwereveryclosetothecut-offCqvaluefortheCobasautomatedreadoutsoftware.
Moreover,16casesamongthese22sam-plesshowedweakmeltingcurvesignalsintheMeltProHPVTest,buttheirmeltingcurvereadout(Rm)valueswerelowerthanthecut-offRmvaluefortheMeltProautomatedreadoutsoftware,whiletheother6casesshowednomelt-ingcurvesignals.
Ontheotherhand,theRmvaluesforthe41samplesdiagnosedaspositivebytheMeltProHPVTestandnegativebytheCobasHPVTestwerealsoveryclosetothecut-offRmvaluefortheMeltProautomatedreadoutsoftware.
Moreover,23casesamongthese41sam-plesshowedlateamplificationsignalsintheCobasHPVTest,buttheirCqvaluesoccurredlaterthanthecut-offCqvaluefortheCobasautomatedreadoutsoftware,whiletheother18casesshowednoamplificationsignals.
Sixdiscrep-antsamplesinfectedbyHPV-16and5discrepantsamplesinfectedbyHPV-18werediagnosedagainusingathirdpartycomparisonmethodbasedontype-specificreal-timePCR,andallofthemwereconfirmedtobeHPV-16positiveorHPV-18positive(TableS1).
Avisualcomparisonofthedetectionresultsobtainedfromthe1647samplesusingtheMeltProHPVTestandtheCobasHPVTestisshowninFig.
1.
Atotalof339sampleswerediagnosedasHPVpositivebytheMeltProHPVTest,including83samplesthatwerediagnosedasinfectedwithHPV-16orHPV-18.
Amongthese339sam-ples,78.
5%(266/339)sampleswereidentifiedashavingasingleHPVtypeinfection,whereas21.
5%(73/339)sam-pleswereinfectedbymultipleHPVtypes.
Bycomparison,atotalof321sampleswerediagnosedasHPVpositivebytheCobasHPVTest,including81samplesthatwerediagnosedwithHPV-16orHPV-18infection.
However,becausetheCobasHPVTestcannotgenotypetheother12high-riskHPVtypes,asidefromHPV-16andHPV-18,wecouldnotcalculatethenumberofsamplesinfectedbymultipleHPVtypes.
Thedistributionof14high-riskHPVtypesamongthese1647samplesisshowninFig.
2.
HPV-52(70cases),HPV-58(64cases),andHPV-16(59cases)werethethreemostprevalentHPVtypesinthisstudy.
HPV-31(8cases)andHPV-45(5cases)werethetworarestHPVtypes.
Thedistri-butionsoftheothertypeswereHPV-39(31cases),HPV-68(29cases),HPV-18(27cases),HPV-51(26cases),HPV-56(23cases),HPV-59(20cases),HPV-33(18cases),HPV-66(17cases),andHPV-35(15cases).
Since21.
5%ofthepositivesampleswereinfectedwithmultipleHPVtypes,thetotalnumberofHPVtypecases(412cases)waslargerthanthetotalnumberofHPVpositivesamples(339samples).
Tworepeatsusingparalleldiagnosisofthe540samplesinanintra-laboratorystudyshowed97.
8%agreement,withakappavalueof0.
947(0.
917-0.
977,95%CI).
Theintra-laboratorydataforeachHPVtypeisshowninTableS2.
Theinter-laboratorystudyof540samplesshowed96.
9%agreement,withakappavalueof0.
925(0.
889-0.
961,95%Table1ComparisonofHPVdetection,HPV-16genotyping,andHPV-18genotypingusingtheMeltProHPVTestandtheCobasHPVTestMeltProHPVCobasHPVTotalKappavalue(95%CI)HPV(+)HPV()HPV(+)300413410.
881(0.
851-0.
911)HPV()2212841306Total32213251647MeltProHPVCobasHPVTotalKappavalue(95%CI)HPV-16(+)HPV-16()HPV-16(+)554590.
946(0.
902-0.
990)HPV-16()215861588Total5715901647MeltProHPVCobasHPVTotalKappavalue(95%CI)HPV-18(+)HPV-18()HPV-18(+)243270.
904(0.
818-0.
990)HPV-18()216181620Total2616211647728Z.
Tangetal.
CI).
Theinter-laboratorydataforeachHPVtypeisshowninTableS3.
Regularscreeningforhigh-riskHPVisrecommendedforadultwomanbytheWHOformonitoringthedevelop-mentofcervicalcancer[3].
Inthisstudy,weevaluatedtheclinicalperformanceoftheMeltProHPVTestusing1647samplescollectedfromwomenduringroutinemedi-calexaminations.
TheHPVdetectionresultsandHPV-16andHPV18genotypingresultsusingtheMeltProHPVTestwereinstrongagreementwithdatafromtheCobasHPVTest(Table1).
AllinconsistentcasesbetweentheMeltProHPVTestandtheCobasHPVTestwerefoundtobesamplesinfectedwithalowviralloadofHPV.
61.
9%(16+23cases/63cases)ofthenegativeresultsforthe63discrepantcasesshowedweakdetectionsignalsbelowthecut-offvaluesfortheMeltProandCobasautomatedreadoutsoftware.
BoththeMeltProHPVTestandtheCobasHPVTestweredesignedforthesimultaneousdetectionof14high-riskHPVtypesinasinglereaction,whichmeansthattheampli-ficationprimersusedcannotcompletelymatchthegenesequenceforeachHPVtype.
Differingamplificationprim-ersmightbeanimportantreasonforthediscrepantresultsobtainedforthese63samplesusingthetwomethods.
HPVtype-specificreal-timePCRwasusedasathirddetectionmethodforthesediscrepantsamplesinfectedbyHPV-16orHPV-18(TableS1).
AthirddetectionmethodwasdesignedtocontaintwopairsofprimersthatperfectlymatchedHPV-16orHPV-18andshouldonlydetectthesetwoHPVtypes.
AsshownintableS1,thedetectionresultsforHPVtype-specificreal-timePCRshowedthat11'suspicious'samplesinfectedbyHPV-16orHPV-18werepositive.
HPVdetectionHPVgenotypingHPVnegative(N=1326)HPVnegative(N=1308)HPVpostive(N=321)HPVpostive(N=339)HPV-16(N=32)HPV-18(N=17)HPV-16or18&othertype(N=32)HPV-16(N=36)HPV-18(N=20)HPV-16or18&othertype(N=27)Othertype(N=242)HPV-52(N=48)HPV-58(N=45)HPV-39(N=20)HPV-51(N=17)HPV-68(N=17)HPV-66(N=15)HPV-56(N=14)HPV-59(N=12)HPV-33(N=10)HPV-31(N=5)HPV-35(N=5)HPV-45(N=2)Co-infection(N=46)CobasHPVtestMeltProHPVtestFig.
1Detectionresultsfor1647samplesusingtheMeltProHPVTestandtheCobasHPVTest.
TheleftpanelshowsthestatisticaldatafortheHPVdetectionresults,prasinouspie:HPVnegativesam-ples,pinkpie:HPVpositivesamples.
Therightpanelshowsthesta-tisticaldatafortheHPVgenotypingresults,redpie:HPV-16,yellowpie:HPV-18,greenpie:co-infectionsamplesincludingHPV-16orHPV-18,bluepie:HPV-othertype(fortheMetltProHPVTest,fromdarktolight:HPV-52,58,39,51,68,66,56,59,33,31,35,and45),greypie:co-infectionsamplesexcludingHPV-16andHPV-181618525839685156593366353145204060HPVNumberofHPVtypeFig.
2ThedistributionoftheHPVgenotypingresultsusingtheMeltProHPVTest.
Redbars:HPV-16andHPV-18,bluebars:otherhigh-riskHPVtypes729ComparedtotheCobasHPVsystem,theMeltProHPVTestprovidesfullgenotypinginformationfor14high-riskHPVtypes.
ThismeanstheMeltProHPVTestcanidentifythespecificHPVtypewithinasampleduringthedetectionstep(Fig.
1),whichwillbenefitusersconductingHPVepi-demiologicalstudies.
Inthisstudy,HPV-16,HPV-52,andHPV-58werethethreemostprevalenthigh-riskHPVtypesinthesoutheastofChina(Fig.
2).
Thisconclusioniscon-sistentwithpreviousreportsbyotherresearchers[19,20].
Consequently,HPV-52andHPV-58shouldbeconsideredforcoverageduringHPVvaccinedevelopmentbyChinesescientists.
Thefullgenotypingofhigh-riskHPVcanalsohelpdoctorsdeterminewhetherapatienthasbeenpersis-tentlyinfectedbythesameHPVtypeorinfectedmultipletimesbydifferentHPVtypes.
Insummary,theMeltProHPVTestandCobasHPVTestarecomparable,with96.
2%agreementandakappacoefficientof0.
881forHPVdetection.
AlthoughtheMeltProHPVTestcanidentifymoreHPVtypesthantheCobasHPVTest,theirclinicalperformancesweresimi-larregardingHPVdetection.
Differentcut-offvaluesandthedifferentamplificationprimersmightbethecauseofdiscrepantresultsbetweenthetwomethods.
Thereproduc-ibilityoftheMeltProHPVTestprovedtobestableinbothintra-laboratoryandinter-laboratorystudies.
TheMeltProHPVTestcanprovidefulltypinginformationandisanaccu-rate,high-throughput,andlow-costmethodthatcanbeusedinthefutureforclinicalHPVscreeningandgenotyping.
AcknowledgementsWethankNationalNaturalScienceFoundation(81401724toY.
Liao),KeyProjectofCooperationProgramforUni-versityandIndustryofFujianProvince(2013Y4008toY.
Xu)andCooperationProgramforUniversityandIndustryofXiamenCity(3502Z20173013toY.
Liao)forfinancialsupport.
CompliancewithethicalstandardsConflictofinterestQ.
LiholdsequityinterestinZeesanBiotech.
Allofotherauthorsdeclarethattheyhavenoconflictofinterest.
TheMeltProHPVtestreagentswerekindlyprovidedbyZeesanBiotech.
Humam/animalrightsstatementThisarticledoesnotcontainanystudieswithhumanparticipantsoranimalsperformedbyanyoftheauthors.
References1.
WoodmanCBJ,CollinsSI,YoungLS(2007)ThenaturalhistoryofcervicalHPVinfection:unresolvedissues.
NatRevCancer7:11–222.
HenryCK,MaribelA,ClaireT,PaulaW,AlexandraS,BoykaS,ClareG,HeleneB,ChristopherR,RobinD,MinaD,JeanM,AndrewB,AndrewT,SueM,JulianP(2009)HPVtestingincombinationwithliquid-basedcytologyinprimarycervicalscreening(ARTISTIC):arandomizedcontrolledtrial.
LancetOncol10:672–6823.
WorldHealthOrganization(2010)Humanpapillomaviruslabo-ratorymanua.
Section5:35–634.
RodenR,WuTC(2006)HowwillHPVvaccinesaffectcervicalcancerNatRevCancer6:753–7635.
MuozN,BoschFX,DeSanjoséS,HerreroR,CastellsaguéX,ShahKV,SnijdersPJF,MeijerCJLM(2003)Epidemiologicclassificationofhumanpapillomavirustypesassociatedwithcervicalcancer.
NEnglJMed.
348:518–5276.
LrinczAT(1996)HybridCapturemethodfordetectionofhumanpapillomavirusDNAinclinicalspecimens:atoolforclinicalmanagementofequivocalPapsmearsandforpopulationscreening.
JObstetGynaecolRes22:629–6367.
GravittP,PeytonC,AppleR,WheelerC(1998)Genotypingof27humanpapillomavirustypesbyusingL1consensusPCRproductsbyasinglehybridization,reverselineblotdetectionmethod.
J.
Clin.
Microbiol36:3020–30278.
KleterB,VanDoornLJ,SchrauwenL,MolijnA,SastrowijotoS,TerScheggetJ,LindemanJ,HarmselBT,BurgerM,QuintW(1999)Developmentandclinicalevaluationofahighlysen-sitivePCR-reversehybridizationlineprobeassayfordetectionandidentificationofanogenitalhumanpapillomavirus.
JClinMicrobiol37:2508–25179.
LiuSS,LeungRCY,ChanKKL,CheungANY,NganHYS(2010)EvaluationofanewlydevelopedGenoArrayhumanpapillomavirus(HPV)genotypingassayandcomparisonwiththeRochelineararrayHPVgenotypingassay.
JClinMicrobiol48:758–76410.
HeidemanD,HesselinkA,BerkhofJ,vanKemenadeF,Melch-ersW,DaalmeijerNF,VerkuijtenM,MeijerCJLM,SnijdersPJF(2011)ClinicalvalidationoftheCobas4800HPVtestforcervicalscreeningpurposes.
JClinMicrobiol49:3983–398511.
CuzickJ,AmbroisineL,CadmanL,AustinJ,HoL,TerryG,Lid-dleS,DinaR,McCarthyJ,BuckleyH,BergeronC,SoutterWP,LyonsD,SzarewskiA(2010)PerformanceoftheAbbottrealtimehigh-riskHPVtestinwomenwithabnormalcervicalcytologysmears.
JMedVirol82:1186–119112.
HwangY,LeeM(2012)ComparisonoftheAdvanSurehumanpapillomavirusscreeningreal-timePCR,theAbbottrealtimehighriskhumanpapillomavirustest,andthehybridcapturehumanpapillomavirusDNAtestforthedetectionofhumanpapilloma-virus.
AnnLabMed32:201–20513.
MicalessiIM,BouletGA,BogersJJ,BenoyIH,DepuydtCE(2012)High-throughputdetection,genotypingandquantificationofthehumanpapillomavirususingreal-timePCR.
ClinChemLabMed50:655–66114.
LiaoY,ZhouY,GuoQ,XieX,LuoE,LiJ,LiQ(2013)Simul-taneousdetection,genotyping,andquantificationofhumanpap-illomavirusesbymulticolorreal-timePCRandmeltingcurveanalysis.
JClinMicrobiol51:429–43515.
ArbynM,DepuydtC,BenoyI,BogersJ,CuschieriK,SchmittM,PawlitaM,GeraetsD,HeardI,GheitT,TommasinoM,PoljakM,BondeJ,QuintW(2016)VALGENT:aprotocolforclinicalvali-dationofhumanpapillomavirusassays.
JClinVirol76:S14–S2116.
GeraetsDT,CuschieriK,deKoningMN,vanDoornLJ,SnijdersPJ,MeijerCJ,QuintWG,ArbynM(2014)ClinicalevaluationofaGP5+/6+-basedluminexassayhavingfullhigh-riskhumanpapillomavirusgenotypingcapabilityandaninternalcontrol.
JClinMicrobiol52:3996–400217.
CuschieriK,GeraetsDT,MooreC,QuintW,DuvallE,ArbynM(2015)ClinicalandanalyticalperformanceoftheOnclarityHPVassayusingtheVALGENTframework.
JClinMicrobiol53:3272–327918.
HeardI,CuschieriK,GeraetsDT,QuintW,ArbynM(2016)ClinicalandanalyticalperformanceofthePapilloCheck730Z.
Tangetal.
HPV-ScreeningassayusingtheVALGENTframework.
JClinVirol81:6–1119.
ShixuanH,IrinaA,BethAM,AnnaMB(1997)Humanpapil-lomavirustypes52and58areprevalentincervicalcancersfromChinesewomen.
IntJCancer70:408–41120.
ShuangL,XiaoC,MinL,FengxiaC,LiangM,YongtongC(2015)Distributionofhigh-riskhumanpapillomavirusgeno-typesinHPV-infectedwomeninBeijing,China.
JMedVirol87:504–507
org/10.
1007/s00705-017-3645-1BRIEFREPORTAcomparisonoftheMeltProHPVTestwiththeCobasHPVTestfordetectingandgenotyping14highriskhumanpapillomavirustypesZhitengTang1·YeXu2·NajieSong3·DongqingZou3·YiqunLiao4·QinggeLi2·ChaoPan1Received:11May2017/Accepted:3October2017/Publishedonline:5December2017Springer-VerlagGmbHAustria,partofSpringerNature2017AbstractTheclinicalperformanceofthenewlydevelopedMeltProHPVTest,basedonmulticolormeltingcurveanalysis,wasevaluatedandcomparedwiththecommerciallyavailableCobasHPVTestfordetectionofHPVandgenotypingofHPV-16andHPV-18.
Atotalof1647cervicalsampleswereanalyzedwithbothtests.
Theagreementvalueswere96.
2%forHPVdetection,99.
6%forHPV-16identification,and99.
7%forHPV-18identification.
AllgenotypingresultsfromMeltProHPVTestshowedthatHPV-52,HPV-58,andHPV-16werethemostcommontypesinthisstudy.
Intra-laboratoryreproducibilitystudiesshowed97.
8%agreementwhileinter-laboratoryreproducibilitystudiesshowed96.
9%agreementfortheMeltProHPVTest.
TheMeltProHPVTestandCobasHPVTestarehighlycorrelativeandareusefulformonitoringHPVinfection.
High-riskhumanpapillomavirus(HPV)infectionisahighriskfactorforthedevelopmentofcervicalcancer,whichisthesecondmostcommonmalignanttumorinwomenworld-wide[1].
High-riskHPVdiagnosescombinedwithliquid-basedcytologyanalysisisconsideredtobethemosteffectivemethodforearlycervicalcancerscreening[2].
Thefirstedi-tionofthe"Humanpapillomaviruslaboratorymanual"waspublishedin2009bytheWorldHealthOrganization(WHO)toprovideguidanceforhigh-riskHPVdiagnoses[3].
HPVvaccinationiscurrentlythesafestapproachforpreventingthedevelopmentofcervicalcancer[4].
ThedistributionofHPVtypeandtheindividualriskofeachHPVtypearetwoimportantfactorsthatneededtobeconsideredfordevelopingtheHPVvaccineagainstparticularHPVgenotypeswithinacountry[5].
Convenientandaccuratetechniquesforhigh-riskHPVdetectionandgenotypingareurgentlyneededforHPVclinicaldiagnosesandepidemiologicalstudies.
ThehybridizationcapturedetectionforHPVgeneticDNAoritsPCRampliconisatraditionalapproachforHPVgeno-typing[6–9].
However,itrequiresmanyoperationalsteps,andPCRcontaminationisacommonproblem.
Avarietyofmethodsbasedonreal-timePCRhavebeendevelopedintheHandlingEditor:ZhongjieShi.
ZhitengTangandYeXucontributedequallytothiswork.
ElectronicsupplementarymaterialTheonlineversionofthisarticle(https://doi.
org/10.
1007/s00705-017-3645-1)containssupplementarymaterial,whichisavailabletoauthorizedusers.
*YiqunLiaoyqliao@xmu.
edu.
cn*QinggeLiqgli@xmu.
edu.
cn*ChaoPanpanchao123a@126.
com1ZhongshanHospital,XiamenUniversity,Xiamen,Fujian,China2TheStateKeyLaboratoryofCellularStressBiology,StateKeyLaboratoryofMolecularVaccinologyandMolecularDiagnostics,EngineeringResearchCenterofMolecularDiagnosticsoftheMinistryofEducation,SchoolofLifeSciences,XiamenUniversity,Xiamen,Fujian,China3ZeesanBiotechnologyCompany,Xiamen,Fujian,China4TheStateKeyLaboratoryofMolecularVaccinologyandMolecularDiagnostics,StateKeyLaboratoryofCellularStressBiology,EngineeringResearchCenterofMolecularDiagnosticsoftheMinistryofEducation,SchoolofPublicHealth,XiamenUniversity,Xiamen,Fujian,China726Z.
Tangetal.
pastdecade[10–13].
Comparedwithtraditionalhybridizationcapturedetection,thereal-timePCRplatformpossessestheadvantagesofconvenience,highthroughput,lowtimeandcost,andlowriskoffalse-positiveresultsduetocross-con-taminationofPCR.
Commercialreal-timePCRkitsarenowwidelyadoptedinclinicalHPVdiagnosticandresearchstud-ies.
TheCobasHPVTestisatypicalsystemthathasbeenapprovedbytheU.
S.
FoodandDrugAdministration(FDA).
InapreviousstudywereportedanovelHPVgenotyp-ingmethodbasedonreal-timePCRandmeltcurveanalysis[14].
TheMeltProHPVTestwasdevelopedwiththesamemethod,andcandetectandgenotypethe14mostcommonhigh-riskHPVtypesinasingle-tubereaction.
Inthisstudy,weperformedacomparisonoftheMeltProHPVTestandtheCobasHPVTestforthedetectionandgenotypingof14high-riskHPVtypesinsoutheastChina.
TheprocessesinvolvedinthiscomparisonstudyareshowninFig.
S1.
More-over,reproducibilityisimportantforanynewlydevelopedmethod.
AprotocolforclinicalvalidationofHPVassays,called"VALGENT",wasdevelopedandsuccessfullyappliedforthecomparisonofmanycommercialHPVtests[15–18].
Followingthisguideline,intra-laboratoryandinter-laboratoryreproducibilityexperimentswereperformedinthisstudy.
ForcomparisonoftheMeltProHPVTestandtheCobasHPVTest,atotalof1647residualcervixcellsam-pleswerecollectedfromindividualwomenlivinginthesoutheastofChina.
Pregnantwomenwereexcludedfromthisstudy.
AllsampleswerecollectedatZhongshanHos-pital,XiamenUniversity(Xiamen,Fujian,China)withaThinPrepliquid-basedcytologysystem,in2015,andstoredat-20°Cfor2weeksbeforeanalysis.
Theageofthepatientsisfrom19to65yearsoldwithameanageof32years-oldandamedianof31years-old.
AlloftheexperimentsinthiscomparisonstudywereperformedatZhongshanHospital.
Fortheintra-laboratoryandinter-laboratoryreproducibilitystudyoftheMeltProHPVTest,atotalof540residualcervixcellsampleswerecollectedatZhongshanhospitalbythesameprotocolin2017.
Theintra-laboratoryreproduc-ibilitystudywasperformedatZhongshanHospital,whiletheinter-laboratoryreproducibilitiesstudywasperformedatZhongshanHospitalandtheEngineeringResearchCenterofMolecularDiagnosticsoftheMinistryofEducation,Xia-menUniversity.
ThestudyprotocolwasapprovedbyTheResearchEthicsCommitteeofXiamenUniversity.
Forthecomparisonstudy,1647sampleswereassayedinadouble-blindedfashionusingtheMeltProHPVTest(ZeesanBiotechnologyCo.
,China)andtheCobasHPVTest(RocheDiagnosticsCo.
,Switzerland)onthesameday.
DNAextrac-tionwasautomaticallyperformed,separately,accordingtotheprotocolsofthesetwosystems.
TheMeltProHPVTestdetected14high-riskHPVtypes(HPV-16,18,31,33,35,39,45,51,52,56,58,59,66,and68)andprovidedspecificgeno-typingforall14HPVtypesbasedonmeltcurveanalysis.
Theglyceraldehyde-3-phosphatedehydrogenase(GAPDH)genewasusedasaninternalcontrolintheMeltProHPVTest.
ThePCRreactionanddataprocessingstepsoftheMeltProHPVTestwereperformedintheSLAN96real-timePCRsystem.
TheCobasHPVTestdetectedthesame14high-riskHPVtypesbutonlyprovidedspecificgenotypingforHPV-16andHPV-18,basedonreal-timePCR.
Theβ-globulingenewasusedasinternalcontrolintheCobasHPVTest.
ThePCRreactionanddataprocessingstepsoftheCobasHPVTestwereperformedinaCobas4800real-timePCRsystem.
TheexperimentalconditionsforTheMeltProHPVTestandCobasHPVTestfollowedtheguidelinesoftheirassociatedprotocols.
Duringeachrun,forbothassays,apositiveandnegativecontrolwasincludedtoensureproperPCRreactionsandthattherewasnocarry-overcontamination.
TheHPVgenotypeforeachsamplewasassayedusingindependent,automated-readoutsoftwaresuppliedwiththeMeltProHPVsystemorCobasHPVsystems.
Thediag-nosticresultsfromtheMeltProHPVTestandtheCobasHPVTestwerecomparedtoevaluateagreement.
Foralldis-crepantsamples,theoriginaldatawereprocessedmanually.
ForcaseswithdiscrepantresultsofHPV-16andHPV-18,confirmatorytestingwasperformedusinganHPVtype-specificreal-timePCRassaydiagnostickitdesignedonlyforHPV-16andHPV-18(KehuaLtd.
,Shanghai,China).
Fortheintra-laboratoryreproducibilitystudy,540sam-pleswereassayedbytheMeltProHPVTesttwiceonthesamedayatZhongshanhospital,andtheresultsofthefirstandsecondassaywerecompared.
Fortheinter-laboratoryreproducibilitystudy,thesame540sampleswereassayedoneweeklaterattheEngineeringResearchCenterofMolecularDiagnosticsoftheMinistryofEducation,andtheresultswerecomparedwiththefirstassay'sresultsfromtheintra-laboratoryreproducibilitystudy.
StatisticalanalysisforthecomparisondatawascarriedoutusingtheSPSSstatisticalsoftware(version13.
0,SPSSInc.
,Chicago,IL).
Epidemiologicalprevalence,forthe14high-riskHPVtypes,wasonlycalculatedfromresultsobtainedwiththeMeltProHPVTest,whichwascapableoffullgenotyping.
Wecomparedtheresultsobtainedfrom1647samplesusingtheMeltProHPVTestandtheCobasHPVTestforthedetectionof14high-riskHPVtypeswithoutgenotyping(Table1).
Theoverallagreementbetweenthesetestswas96.
2%(1584/1647),andthekappavaluewas0.
881(95%CI,0.
851-0.
911).
SincetheMeltProHPVTestandtheCobasHPVTestbothprovideHPV-16andHPV-18geno-typing,wealsoevaluatedthegenotypingresultsforHPV-16andHPV-18,withinthecomparisonshowninTable1.
TheagreementforthegenotypingresultsforHPV-16was99.
6%(1641/1647),andthekappavaluewas0.
946(95%CI,0.
902-0.
990).
TheagreementforthegenotypingresultsforHPV-18was99.
7%(1642/1647),andthekappavaluewas7270.
904(95%CI,0.
818-0.
990).
TheresultsfortheHPVdetec-tion,HPV-16genotypingandHPV-18genotypingusingtheMeltProHPVTestandtheCobasHPVTestwerenotsignificantlydifferent(McNemar'sTest,Pvalue=0.
23forHPVdetection,Pvalue=0.
69forHPV-16genotyping,Pvalue=1.
00forHPV-18genotyping).
Forthe63discrepantsamples,weprocessedtheorigi-naldatamanually.
ThequantitativePCRcycle(Cq)valuesforthe22samplesdiagnosedaspositivebytheCobasHPVTestandnegativebytheMeltProHPVTestwereveryclosetothecut-offCqvaluefortheCobasautomatedreadoutsoftware.
Moreover,16casesamongthese22sam-plesshowedweakmeltingcurvesignalsintheMeltProHPVTest,buttheirmeltingcurvereadout(Rm)valueswerelowerthanthecut-offRmvaluefortheMeltProautomatedreadoutsoftware,whiletheother6casesshowednomelt-ingcurvesignals.
Ontheotherhand,theRmvaluesforthe41samplesdiagnosedaspositivebytheMeltProHPVTestandnegativebytheCobasHPVTestwerealsoveryclosetothecut-offRmvaluefortheMeltProautomatedreadoutsoftware.
Moreover,23casesamongthese41sam-plesshowedlateamplificationsignalsintheCobasHPVTest,buttheirCqvaluesoccurredlaterthanthecut-offCqvaluefortheCobasautomatedreadoutsoftware,whiletheother18casesshowednoamplificationsignals.
Sixdiscrep-antsamplesinfectedbyHPV-16and5discrepantsamplesinfectedbyHPV-18werediagnosedagainusingathirdpartycomparisonmethodbasedontype-specificreal-timePCR,andallofthemwereconfirmedtobeHPV-16positiveorHPV-18positive(TableS1).
Avisualcomparisonofthedetectionresultsobtainedfromthe1647samplesusingtheMeltProHPVTestandtheCobasHPVTestisshowninFig.
1.
Atotalof339sampleswerediagnosedasHPVpositivebytheMeltProHPVTest,including83samplesthatwerediagnosedasinfectedwithHPV-16orHPV-18.
Amongthese339sam-ples,78.
5%(266/339)sampleswereidentifiedashavingasingleHPVtypeinfection,whereas21.
5%(73/339)sam-pleswereinfectedbymultipleHPVtypes.
Bycomparison,atotalof321sampleswerediagnosedasHPVpositivebytheCobasHPVTest,including81samplesthatwerediagnosedwithHPV-16orHPV-18infection.
However,becausetheCobasHPVTestcannotgenotypetheother12high-riskHPVtypes,asidefromHPV-16andHPV-18,wecouldnotcalculatethenumberofsamplesinfectedbymultipleHPVtypes.
Thedistributionof14high-riskHPVtypesamongthese1647samplesisshowninFig.
2.
HPV-52(70cases),HPV-58(64cases),andHPV-16(59cases)werethethreemostprevalentHPVtypesinthisstudy.
HPV-31(8cases)andHPV-45(5cases)werethetworarestHPVtypes.
Thedistri-butionsoftheothertypeswereHPV-39(31cases),HPV-68(29cases),HPV-18(27cases),HPV-51(26cases),HPV-56(23cases),HPV-59(20cases),HPV-33(18cases),HPV-66(17cases),andHPV-35(15cases).
Since21.
5%ofthepositivesampleswereinfectedwithmultipleHPVtypes,thetotalnumberofHPVtypecases(412cases)waslargerthanthetotalnumberofHPVpositivesamples(339samples).
Tworepeatsusingparalleldiagnosisofthe540samplesinanintra-laboratorystudyshowed97.
8%agreement,withakappavalueof0.
947(0.
917-0.
977,95%CI).
Theintra-laboratorydataforeachHPVtypeisshowninTableS2.
Theinter-laboratorystudyof540samplesshowed96.
9%agreement,withakappavalueof0.
925(0.
889-0.
961,95%Table1ComparisonofHPVdetection,HPV-16genotyping,andHPV-18genotypingusingtheMeltProHPVTestandtheCobasHPVTestMeltProHPVCobasHPVTotalKappavalue(95%CI)HPV(+)HPV()HPV(+)300413410.
881(0.
851-0.
911)HPV()2212841306Total32213251647MeltProHPVCobasHPVTotalKappavalue(95%CI)HPV-16(+)HPV-16()HPV-16(+)554590.
946(0.
902-0.
990)HPV-16()215861588Total5715901647MeltProHPVCobasHPVTotalKappavalue(95%CI)HPV-18(+)HPV-18()HPV-18(+)243270.
904(0.
818-0.
990)HPV-18()216181620Total2616211647728Z.
Tangetal.
CI).
Theinter-laboratorydataforeachHPVtypeisshowninTableS3.
Regularscreeningforhigh-riskHPVisrecommendedforadultwomanbytheWHOformonitoringthedevelop-mentofcervicalcancer[3].
Inthisstudy,weevaluatedtheclinicalperformanceoftheMeltProHPVTestusing1647samplescollectedfromwomenduringroutinemedi-calexaminations.
TheHPVdetectionresultsandHPV-16andHPV18genotypingresultsusingtheMeltProHPVTestwereinstrongagreementwithdatafromtheCobasHPVTest(Table1).
AllinconsistentcasesbetweentheMeltProHPVTestandtheCobasHPVTestwerefoundtobesamplesinfectedwithalowviralloadofHPV.
61.
9%(16+23cases/63cases)ofthenegativeresultsforthe63discrepantcasesshowedweakdetectionsignalsbelowthecut-offvaluesfortheMeltProandCobasautomatedreadoutsoftware.
BoththeMeltProHPVTestandtheCobasHPVTestweredesignedforthesimultaneousdetectionof14high-riskHPVtypesinasinglereaction,whichmeansthattheampli-ficationprimersusedcannotcompletelymatchthegenesequenceforeachHPVtype.
Differingamplificationprim-ersmightbeanimportantreasonforthediscrepantresultsobtainedforthese63samplesusingthetwomethods.
HPVtype-specificreal-timePCRwasusedasathirddetectionmethodforthesediscrepantsamplesinfectedbyHPV-16orHPV-18(TableS1).
AthirddetectionmethodwasdesignedtocontaintwopairsofprimersthatperfectlymatchedHPV-16orHPV-18andshouldonlydetectthesetwoHPVtypes.
AsshownintableS1,thedetectionresultsforHPVtype-specificreal-timePCRshowedthat11'suspicious'samplesinfectedbyHPV-16orHPV-18werepositive.
HPVdetectionHPVgenotypingHPVnegative(N=1326)HPVnegative(N=1308)HPVpostive(N=321)HPVpostive(N=339)HPV-16(N=32)HPV-18(N=17)HPV-16or18&othertype(N=32)HPV-16(N=36)HPV-18(N=20)HPV-16or18&othertype(N=27)Othertype(N=242)HPV-52(N=48)HPV-58(N=45)HPV-39(N=20)HPV-51(N=17)HPV-68(N=17)HPV-66(N=15)HPV-56(N=14)HPV-59(N=12)HPV-33(N=10)HPV-31(N=5)HPV-35(N=5)HPV-45(N=2)Co-infection(N=46)CobasHPVtestMeltProHPVtestFig.
1Detectionresultsfor1647samplesusingtheMeltProHPVTestandtheCobasHPVTest.
TheleftpanelshowsthestatisticaldatafortheHPVdetectionresults,prasinouspie:HPVnegativesam-ples,pinkpie:HPVpositivesamples.
Therightpanelshowsthesta-tisticaldatafortheHPVgenotypingresults,redpie:HPV-16,yellowpie:HPV-18,greenpie:co-infectionsamplesincludingHPV-16orHPV-18,bluepie:HPV-othertype(fortheMetltProHPVTest,fromdarktolight:HPV-52,58,39,51,68,66,56,59,33,31,35,and45),greypie:co-infectionsamplesexcludingHPV-16andHPV-181618525839685156593366353145204060HPVNumberofHPVtypeFig.
2ThedistributionoftheHPVgenotypingresultsusingtheMeltProHPVTest.
Redbars:HPV-16andHPV-18,bluebars:otherhigh-riskHPVtypes729ComparedtotheCobasHPVsystem,theMeltProHPVTestprovidesfullgenotypinginformationfor14high-riskHPVtypes.
ThismeanstheMeltProHPVTestcanidentifythespecificHPVtypewithinasampleduringthedetectionstep(Fig.
1),whichwillbenefitusersconductingHPVepi-demiologicalstudies.
Inthisstudy,HPV-16,HPV-52,andHPV-58werethethreemostprevalenthigh-riskHPVtypesinthesoutheastofChina(Fig.
2).
Thisconclusioniscon-sistentwithpreviousreportsbyotherresearchers[19,20].
Consequently,HPV-52andHPV-58shouldbeconsideredforcoverageduringHPVvaccinedevelopmentbyChinesescientists.
Thefullgenotypingofhigh-riskHPVcanalsohelpdoctorsdeterminewhetherapatienthasbeenpersis-tentlyinfectedbythesameHPVtypeorinfectedmultipletimesbydifferentHPVtypes.
Insummary,theMeltProHPVTestandCobasHPVTestarecomparable,with96.
2%agreementandakappacoefficientof0.
881forHPVdetection.
AlthoughtheMeltProHPVTestcanidentifymoreHPVtypesthantheCobasHPVTest,theirclinicalperformancesweresimi-larregardingHPVdetection.
Differentcut-offvaluesandthedifferentamplificationprimersmightbethecauseofdiscrepantresultsbetweenthetwomethods.
Thereproduc-ibilityoftheMeltProHPVTestprovedtobestableinbothintra-laboratoryandinter-laboratorystudies.
TheMeltProHPVTestcanprovidefulltypinginformationandisanaccu-rate,high-throughput,andlow-costmethodthatcanbeusedinthefutureforclinicalHPVscreeningandgenotyping.
AcknowledgementsWethankNationalNaturalScienceFoundation(81401724toY.
Liao),KeyProjectofCooperationProgramforUni-versityandIndustryofFujianProvince(2013Y4008toY.
Xu)andCooperationProgramforUniversityandIndustryofXiamenCity(3502Z20173013toY.
Liao)forfinancialsupport.
CompliancewithethicalstandardsConflictofinterestQ.
LiholdsequityinterestinZeesanBiotech.
Allofotherauthorsdeclarethattheyhavenoconflictofinterest.
TheMeltProHPVtestreagentswerekindlyprovidedbyZeesanBiotech.
Humam/animalrightsstatementThisarticledoesnotcontainanystudieswithhumanparticipantsoranimalsperformedbyanyoftheauthors.
References1.
WoodmanCBJ,CollinsSI,YoungLS(2007)ThenaturalhistoryofcervicalHPVinfection:unresolvedissues.
NatRevCancer7:11–222.
HenryCK,MaribelA,ClaireT,PaulaW,AlexandraS,BoykaS,ClareG,HeleneB,ChristopherR,RobinD,MinaD,JeanM,AndrewB,AndrewT,SueM,JulianP(2009)HPVtestingincombinationwithliquid-basedcytologyinprimarycervicalscreening(ARTISTIC):arandomizedcontrolledtrial.
LancetOncol10:672–6823.
WorldHealthOrganization(2010)Humanpapillomaviruslabo-ratorymanua.
Section5:35–634.
RodenR,WuTC(2006)HowwillHPVvaccinesaffectcervicalcancerNatRevCancer6:753–7635.
MuozN,BoschFX,DeSanjoséS,HerreroR,CastellsaguéX,ShahKV,SnijdersPJF,MeijerCJLM(2003)Epidemiologicclassificationofhumanpapillomavirustypesassociatedwithcervicalcancer.
NEnglJMed.
348:518–5276.
LrinczAT(1996)HybridCapturemethodfordetectionofhumanpapillomavirusDNAinclinicalspecimens:atoolforclinicalmanagementofequivocalPapsmearsandforpopulationscreening.
JObstetGynaecolRes22:629–6367.
GravittP,PeytonC,AppleR,WheelerC(1998)Genotypingof27humanpapillomavirustypesbyusingL1consensusPCRproductsbyasinglehybridization,reverselineblotdetectionmethod.
J.
Clin.
Microbiol36:3020–30278.
KleterB,VanDoornLJ,SchrauwenL,MolijnA,SastrowijotoS,TerScheggetJ,LindemanJ,HarmselBT,BurgerM,QuintW(1999)Developmentandclinicalevaluationofahighlysen-sitivePCR-reversehybridizationlineprobeassayfordetectionandidentificationofanogenitalhumanpapillomavirus.
JClinMicrobiol37:2508–25179.
LiuSS,LeungRCY,ChanKKL,CheungANY,NganHYS(2010)EvaluationofanewlydevelopedGenoArrayhumanpapillomavirus(HPV)genotypingassayandcomparisonwiththeRochelineararrayHPVgenotypingassay.
JClinMicrobiol48:758–76410.
HeidemanD,HesselinkA,BerkhofJ,vanKemenadeF,Melch-ersW,DaalmeijerNF,VerkuijtenM,MeijerCJLM,SnijdersPJF(2011)ClinicalvalidationoftheCobas4800HPVtestforcervicalscreeningpurposes.
JClinMicrobiol49:3983–398511.
CuzickJ,AmbroisineL,CadmanL,AustinJ,HoL,TerryG,Lid-dleS,DinaR,McCarthyJ,BuckleyH,BergeronC,SoutterWP,LyonsD,SzarewskiA(2010)PerformanceoftheAbbottrealtimehigh-riskHPVtestinwomenwithabnormalcervicalcytologysmears.
JMedVirol82:1186–119112.
HwangY,LeeM(2012)ComparisonoftheAdvanSurehumanpapillomavirusscreeningreal-timePCR,theAbbottrealtimehighriskhumanpapillomavirustest,andthehybridcapturehumanpapillomavirusDNAtestforthedetectionofhumanpapilloma-virus.
AnnLabMed32:201–20513.
MicalessiIM,BouletGA,BogersJJ,BenoyIH,DepuydtCE(2012)High-throughputdetection,genotypingandquantificationofthehumanpapillomavirususingreal-timePCR.
ClinChemLabMed50:655–66114.
LiaoY,ZhouY,GuoQ,XieX,LuoE,LiJ,LiQ(2013)Simul-taneousdetection,genotyping,andquantificationofhumanpap-illomavirusesbymulticolorreal-timePCRandmeltingcurveanalysis.
JClinMicrobiol51:429–43515.
ArbynM,DepuydtC,BenoyI,BogersJ,CuschieriK,SchmittM,PawlitaM,GeraetsD,HeardI,GheitT,TommasinoM,PoljakM,BondeJ,QuintW(2016)VALGENT:aprotocolforclinicalvali-dationofhumanpapillomavirusassays.
JClinVirol76:S14–S2116.
GeraetsDT,CuschieriK,deKoningMN,vanDoornLJ,SnijdersPJ,MeijerCJ,QuintWG,ArbynM(2014)ClinicalevaluationofaGP5+/6+-basedluminexassayhavingfullhigh-riskhumanpapillomavirusgenotypingcapabilityandaninternalcontrol.
JClinMicrobiol52:3996–400217.
CuschieriK,GeraetsDT,MooreC,QuintW,DuvallE,ArbynM(2015)ClinicalandanalyticalperformanceoftheOnclarityHPVassayusingtheVALGENTframework.
JClinMicrobiol53:3272–327918.
HeardI,CuschieriK,GeraetsDT,QuintW,ArbynM(2016)ClinicalandanalyticalperformanceofthePapilloCheck730Z.
Tangetal.
HPV-ScreeningassayusingtheVALGENTframework.
JClinVirol81:6–1119.
ShixuanH,IrinaA,BethAM,AnnaMB(1997)Humanpapil-lomavirustypes52and58areprevalentincervicalcancersfromChinesewomen.
IntJCancer70:408–41120.
ShuangL,XiaoC,MinL,FengxiaC,LiangM,YongtongC(2015)Distributionofhigh-riskhumanpapillomavirusgeno-typesinHPV-infectedwomeninBeijing,China.
JMedVirol87:504–507
imidc:$88/月,e3-1230/16G内存/512gSSD/30M直连带宽/13个IPv4日本多IP
imidc对日本独立服务器在搞特别促销,原价159美元的机器现在只需要88美元,而且给13个独立IPv4,30Mbps直连带宽,不限制流量。注意,本次促销只有一个链接,有2个不同的优惠码,你用不同的优惠码就对应着不同的配置,价格也不一样。88美元的机器,下单后默认不管就给512G SSD,要指定用HDD那就发工单,如果需要多加一个/28(13个)IPv4,每个月32美元...官方网站:https:...
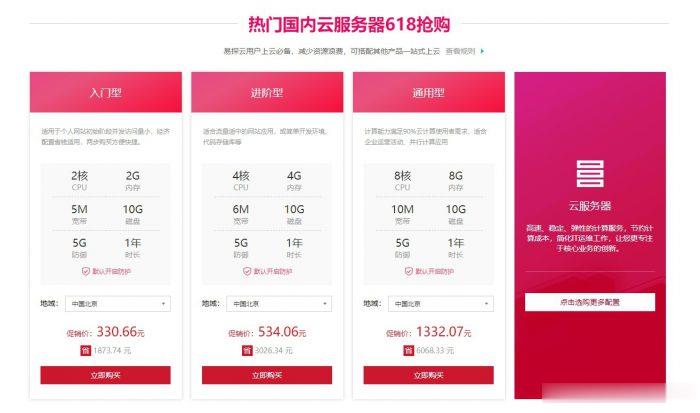
易探云2核2G5M仅330元/年起,国内挂机宝云服务器,独立ip
易探云怎么样?易探云是国内一家云计算服务商家,致力香港服务器、国内外服务器租用及托管等互联网业务,目前主要地区为运作香港BGP、香港CN2、广东、北京、深圳等地区。目前,易探云推出深圳或北京地区的适合挂机和建站的云服务器,国内挂机宝云服务器(可选深圳或北京地区),独立ip;2核2G5M挂机云服务器仅330元/年起!点击进入:易探云官方网站地址易探云国内挂机宝云服务器推荐:1、国内入门型挂机云服务器...
EdgeNat 新年开通优惠 - 韩国独立服务器原生IP地址CN2线路七折优惠
EdgeNat 商家在之前也有分享过几次活动,主要提供香港和韩国的VPS主机,分别在沙田和首尔LG机房,服务器均为自营硬件,电信CN2线路,移动联通BGP直连,其中VPS主机基于KVM架构,宿主机采用四路E5处理器、raid10+BBU固态硬盘!最高可以提供500Gbps DDoS防御。这次开年活动中有提供七折优惠的韩国独立服务器,原生IP地址CN2线路。第一、优惠券活动EdgeNat优惠码(限月...

najie为你推荐
-
支付宝查询余额支付宝里如何查询银行卡里面的余额?缓冲区溢出教程哪里可以下载黑客教程,详细网址,淘宝店推广如何推广淘宝店申请证书申请毕业证书苹果5怎么越狱苹果5怎么越狱?qq怎么发邮件如何通过QQ发送邮件数据库损坏数据库坏了怎么办lockdownd[求教]在淘宝买了张激活卡,请问怎么取消激活网站地图制作我想给网站做网站地图不知道怎么做的,请教高手!cisco防火墙cisco防火墙里k9是什么意思