Liguejusewang
jusewang 时间:2021-02-23 阅读:()
MicroRNARelatedPolymorphismsandBreastCancerRiskSofiaKhan1,DarioGreco1,2,KyriakiMichailidou3,RogerL.
Milne4,5,TaruA.
Muranen1,TuomasHeikkinen1,KirsimariAaltonen1,6,7,JoeDennis3,ManjeetK.
Bolla3,JianjunLiu8,PerHall9,AstridIrwanto8,KeithHumphreys9,JingmeiLi8,KamilaCzene9,JennyChang-Claude10,RebeccaHein10,11,AnjaRudolph10,PetraSeibold10,DieterFlesch-Janys12,OliviaFletcher13,JulianPeto14,IsabeldosSantosSilva14,NicholaJohnson13,LornaGibson14,ZoeAitken14,JohnL.
Hopper15,HelenTsimiklis16,MinhBui15,EnesMakalic15,DanielF.
Schmidt15,MelissaC.
Southey16,CarmelApicella15,JenniferStone15,QuintenWaisfisz17,HanneMeijers-Heijboer17,MurielA.
Adank17,RobB.
vanderLuijt18,AlfonsMeindl19,RitaK.
Schmutzler20,21,22,23,BertramMu¨ller-Myhsok24,PeterLichtner25,ClareTurnbull26,NazneenRahman26,StephenJ.
Chanock27,DavidJ.
Hunter28,29,AngelaCox30,SimonS.
Cross31,MalcolmW.
R.
Reed30,MarjankaK.
Schmidt32,AnnegienBroeks32,LauraJ.
Van'tVeer32,FransB.
Hogervorst32,PeterA.
Fasching33,34,MichaelG.
Schrauder33,ArifB.
Ekici35,MatthiasW.
Beckmann33,StigE.
Bojesen36,37,BrgeG.
Nordestgaard36,37,SuneF.
Nielsen36,37,HenrikFlyger38,JavierBenitez39,40,PilarM.
Zamora41,JoseI.
A.
Perez42,ChristopherA.
Haiman43,BrianE.
Henderson43,FredrickSchumacher43,LoicLeMarchand44,PaulD.
P.
Pharoah3,45,AlisonM.
Dunning45,MitulShah45,RobertLuben46,JudithBrown3,FergusJ.
Couch47,XianshuWang47,CelineVachon48,JanetE.
Olson48,DietherLambrechts49,50,MatthieuMoisse49,50,RobertParidaens51,Marie-RoseChristiaens51,PascalGuenel52,53,There`seTruong52,53,PierreLaurent-Puig54,ClaireMulot54,FrederickMarme55,56,BarbaraBurwinkel55,57,AndreasSchneeweiss55,56,ChristofSohn55,ElinorJ.
Sawyer58,IanTomlinson59,MichaelJ.
Kerin60,NicolaMiller60,IreneL.
Andrulis61,62,JuliaA.
Knight63,64,SandrineTchatchou61,AnnaMarieMulligan65,66,ThiloDo¨rk67,NataliaV.
Bogdanova68,NataliaN.
Antonenkova69,HodaAnton-Culver70,HatefDarabi9,MikaelEriksson9,MontserratGarcia-Closas71,72,JonineFigueroa27,JolantaLissowska73,LouiseBrinton27,PeterDevilee74,RobertA.
E.
M.
Tollenaar75,CarolineSeynaeve76,ChristiJ.
vanAsperen77,VesselaN.
Kristensen78,79,80,kConFabInvestigators81",AustralianOvarianCancerStudyGroup81,82",SusanSlager48,AmandaE.
Toland83,ChristineB.
Ambrosone84,DrakoulisYannoukakos85,AnnikaLindblom86,SaraMargolin87,PaoloRadice88,PaoloPeterlongo89,MonicaBarile90,PaoloMariani89,91,MaartjeJ.
Hooning92,JohnW.
M.
Martens92,J.
MargrietCollee93,AgnesJager92,AnnaJakubowska94,JanLubinski94,KatarzynaJaworska-Bieniek94,95,KatarzynaDurda94,GrahamG.
Giles4,5,CatrionaMcLean96,HiltrudBrauch97,98,ThomasBru¨ning99,Yon-DschunKo100,TheGENICANetwork97,98,99,100,101,102,103",HermannBrenner104,105,AidaKarinaDieffenbach104,105,VolkerArndt104,ChristaStegmaier106,AnthonySwerdlow107,AlanAshworth13,NickOrr13,MichaelJones71,JacquesSimard108,MarkS.
Goldberg109,110,FranceLabre`che111,MartineDumont108,RobertWinqvist112,KatriPylka¨s112,ArjaJukkola-Vuorinen113,MerviGrip114,VesaKataja115,116,Veli-MattiKosma117,118,119,JaanaM.
Hartikainen117,118,119,ArtoMannermaa117,118,119,UteHamann101,GeorgiaChenevix-Trench120,CarlBlomqvist7,KristiinaAittoma¨ki6,DouglasF.
Easton3,45,HeliNevanlinna1*1DepartmentofObstetricsandGynecology,UniversityofHelsinkiandHelsinkiUniversityCentralHospital,Helsinki,Finland,2FinnishInstituteofOccupationalHealth,Helsinki,Finland,3CentreforCancerGeneticEpidemiology,DepartmentofPublicHealthandPrimaryCare,UniversityofCambridge,Cambridge,UnitedKingdom,4CancerEpidemiologyCentre,CancerCouncilVictoria,Melbourne,Australia,5CentreforEpidemiologyandBiostatistics,MelbourneSchoolofPopulationandGlobalHealth,TheUniversityofMelbourne,Melbourne,Australia,6DepartmentofClinicalGenetics,UniversityofHelsinkiandHelsinkiUniversityCentralHospital,Helsinki,Finland,7DepartmentofOncology,UniversityofHelsinkiandHelsinkiUniversityCentralHospital,Helsinki,Finland,8HumanGeneticsDivision,GenomeInstituteofSingapore,Singapore,Singapore,9DepartmentofMedicalEpidemiologyandBiostatistics,KarolinskaInstitutet,Stockholm,Sweden,10DivisionofCancerEpidemiology,GermanCancerResearchCenter(DKFZ),Heidelberg,Germany,11PMVResearchGroupattheDepartmentofChildandAdolescentPsychiatryandPsychotherapy,UniversityofCologne,Cologne,Germany,12DepartmentofCancerEpidemiology/ClinicalCancerRegistryandInstituteforMedicalBiometricsandEpidemiology,UniversityClinicHamburg-Eppendorf,Hamburg,Germany,13BreakthroughBreastCancerResearchCentre,TheInstituteofCancerResearch,London,UnitedKingdom,14DepartmentofNon-CommunicableDiseaseEpidemiologyDepartment,LondonSchoolofHygieneandTropicalMedicine,London,UnitedKingdom,15CentreforPLOSONE|www.
plosone.
org1November2014|Volume9|Issue11|e109973EpidemiologyandBiostatistics,MelbourneSchoolofPopulationandGlobalHealth,TheUniversityofMelbourne,Melbourne,Australia,16DepartmentofPathology,TheUniversityofMelbourne,Melbourne,Australia,17DepartmentofClinicalGenetics,VUUniversityMedicalCenter,Amsterdam,TheNetherlands,18DepartmentofMedicalGenetics,UniversityMedicalCenterUtrecht,Utrecht,TheNetherlands,19DivisionofGynaecologyandObstetrics,TechnischeUniversita¨tMu¨nchen,Munich,Germany,20DivisionofMolecularGyneco-Oncology,DepartmentofGynaecologyandObstetrics,UniversityHospitalofCologne,Cologne,Germany,21CenterofFamilialBreastandOvarianCancer,UniversityHospitalofCologne,Cologne,Germany,22CenterforIntegratedOncology(CIO),UniversityHospitalofCologne,Cologne,Germany,23CenterforMolecularMedicineCologne(CMMC),UniversityofCologne,Cologne,Germany,24MaxPlanckInstituteofPsychiatry,Munich,Germany,25InstituteofHumanGenetics,HelmholtzZentrumMu¨nchen,GermanResearchCenterforEnvironmentalHealth,Neuherberg,Germany,26SectionofCancerGenetics,InstituteofCancerResearch,Sutton,UnitedKingdom,27DivisionofCancerEpidemiologyandGenetics,NationalCancerInstitute,Rockville,Maryland,UnitedStatesofAmerica,28PrograminMolecularandGeneticEpidemiology,HarvardSchoolofPublicHealth,Boston,Massachusetts,UnitedStatesofAmerica,29DepartmentofEpidemiology,HarvardSchoolofPublicHealth,Boston,Massachusetts,UnitedStatesofAmerica,30CRUK/YCRSheffieldCancerResearchCentre,DepartmentofOncology,UniversityofSheffield,Sheffield,UnitedKingdom,31AcademicUnitofPathology,DepartmentofNeuroscience,UniversityofSheffield,Sheffield,UnitedKingdom,32NetherlandsCancerInstitute,AntonivanLeeuwenhoekhospital,Amsterdam,TheNetherlands,33UniversityBreastCenterFranconia,DepartmentofGynecologyandObstetrics,UniversityHospitalErlangen,Friedrich-AlexanderUniversityErlangen-Nuremberg,ComprehensiveCancerCancerErlangen-EMN,Erlangen,Germany,34DavidGeffenSchoolofMedicine,DepartmentofMedicineDivisionofHematologyandOncology,UniversityofCaliforniaLosAngeles,California,UnitedStatesofAmerica,35InstituteofHumanGenetics,UniversityHospitalErlangen,Friedrich-AlexanderUniversityErlangen-Nuremberg,ComprehensiveCancerCenterErlangen-EMN,Erlangen,Germany,36CopenhagenGeneralPopulationStudy,HerlevHospital,CopenhagenUniversityHospital,Copenhagen,Denmark,37DepartmentofClinicalBiochemistry,HerlevHospital,CopenhagenUniversityHospital,Copenhagen,Denmark,38DepartmentofBreastSurgery,HerlevHospital,CopenhagenUniversityHospital,Copenhagen,Denmark,39HumanGeneticsGroup,HumanCancerGeneticsProgram,SpanishNationalCancerResearchCentre(CNIO),Madrid,Spain,40CentrodeInvestigacionenReddeEnfermedadesRaras(CIBERER),Valencia,Spain,41ServiciodeOncologaMedica,HospitalUniversitarioLaPaz,Madrid,Spain,42ServiciodeCirugaGeneralyEspecialidades,HospitalMonteNaranco,Oviedo,Spain,43DepartmentofPreventiveMedicine,KeckSchoolofMedicine,UniversityofSouthernCalifornia,LosAngeles,California,UnitedStatesofAmerica,44EpidemiologyProgram,CancerResearchCenter,UniversityofHawaii,Honolulu,Hawaii,UnitedStatesofAmerica,45CentreforCancerGeneticEpidemiology,DepartmentofOncology,UniversityofCambridge,Cambridge,UnitedKingdom,46ClinicalGerontology,DepartmentofPublicHealthandPrimaryCare,UniversityofCambridge,Cambridge,UnitedKingdom,47DepartmentofLaboratoryMedicineandPathology,MayoClinic,Rochester,Minnesota,UnitedStatesofAmerica,48DepartmentofHealthSciencesResearch,MayoClinic,Rochester,Minnesota,UnitedStatesofAmerica,49VesaliusResearchCenter(VRC),VIB,Leuven,Belgium,50LaboratoryforTranslationalGenetics,DepartmentofOncology,UniversityofLeuven,Leuven,Belgium,51OncologyDepartment,UniversityHospitalGasthuisberg,Leuven,Belgium,52Inserm(NationalInstituteofHealthandMedicalResearch),CESP(CenterforResearchinEpidemiologyandPopulationHealth),U1018,EnvironmentalEpidemiologyofCancer,Villejuif,France,53UniversityParis-Sud,UMRS1018,Villejuif,France,54UniversiteParisSorbonneCite,UMR-S775Inserm,Paris,France,55DepartmentofObstetricsandGynecology,UniversityofHeidelberg,Heidelberg,Germany,56NationalCenterforTumorDiseases,UniversityofHeidelberg,Heidelberg,Germany,57MolecularEpidemiologyGroup,GermanCancerResearchCenter(DKFZ),Heidelberg,Germany,58ResearchOncology,DivisionofCancerStudies,King'sCollegeLondon,Guy'sHospital,London,UnitedKingdom,59WellcomeTrustCentreforHumanGeneticsandOxfordBiomedicalResearchCentre,UniversityofOxford,Oxford,UnitedKingdom,60ClinicalScienceInstitute,UniversityHospitalGalway,Galway,Ireland,61Lunenfeld-TanenbaumResearchInstituteofMountSinaiHospital,Toronto,Ontario,Canada,62DepartmentofMolecularGenetics,UniversityofToronto,Toronto,Ontario,Canada,63ProssermanCentreforHealthResearch,Lunenfeld-TanenbaumResearchInstitute,MountSinaiHospital,Toronto,Ontario,Canada,64DivisionofEpidemiology,DallaLanaSchoolofPublicHealth,UniversityofToronto,Toronto,Ontario,Canada,65DepartmentofLaboratoryMedicineandPathobiology,UniversityofToronto,Toronto,Ontario,Canada,66DepartmentofLaboratoryMedicine,andtheKeenanResearchCentreoftheLiKaShingKnowledgeInstitute,StMichael'sHospital,Toronto,Ontario,Canada,67DepartmentofObstetricsandGynaecology,HannoverMedicalSchool,Hannover,Germany,68DepartmentofRadiationOncology,HannoverMedicalSchool,Hannover,Germany,69N.
N.
AlexandrovResearchInstituteofOncologyandMedicalRadiology,Minsk,Belarus,70DepartmentofEpidemiology,UniversityofCaliforniaIrvine,Irvine,California,UnitedStatesofAmerica,71DivisionofGeneticsandEpidemiology,InstituteofCancerResearch,Sutton,UnitedKingdom,72BreakthroughBreastCancerResearchCentre,DivisionofBreastCancerResearch,TheInstituteofCancerResearch,London,UnitedKingdom,73DepartmentofCancerEpidemiologyandPrevention,M.
Sklodowska-CurieMemorialCancerCenterandInstituteofOncology,Warsaw,Poland,74DepartmentofHumanGenetics&DepartmentofPathology,LeidenUniversityMedicalCenter,Leiden,Netherlands,75DepartmentofSurgicalOncology,LeidenUniversityMedicalCenter,Leiden,Netherlands,76FamilyCancerClinic,DepartmentofMedicalOncology,ErasmusMC-DanieldenHoedCancerCenter,Rotterdam,Netherlands,77DepartmentofClinicalGenetics,LeidenUniversityMedicalCenter,Leiden,Netherlands,78InstituteofClinicalMedicine,FacultyofMedicine,UniversityofOslo,Oslo,Norway,79DepartmentofClinicalMolecularBiology(EpiGen),UniversityofOslo,Oslo,Norway,80DepartmentofGenetics,InstituteforCancerResearch,OsloUniversityHospital,Radiumhospitalet,Oslo,Norway,81PeterMacCallumCancerCenter,Melbourne,Australia,82QIMRBerghoferMedicalResearchInstitute,Brisbane,Australia,83DepartmentofMolecularVirology,ImmunologyandMedicalGenetics,ComprehensiveCancerCenter,TheOhioStateUniversity,Columbus,Ohio,UnitedStatesofAmerica,84RoswellParkCancerInstitute,Buffalo,NewYork,UnitedStatesofAmerica,85MolecularDiagnosticsLaboratory,IRRP,NationalCentreforScientificResearch"Demokritos",AghiaParaskeviAttikis,Athens,Greece,86DepartmentofMolecularMedicineandSurgery,KarolinskaInstitutet,Stockholm,Sweden,87DepartmentofOncology-Pathology,KarolinskaInstitutet,Stockholm,Sweden,88UnitofMolecularBasesofGeneticRiskandGeneticTesting,DepartmentofPreventiveandPredictiveMedicine,FondazioneIRCCSIstitutoNazionaledeiTumori(INT),Milan,Italy,89IFOM,FondazioneIstitutoFIRCdiOncologiaMolecolare,Milan,Italy,90DivisionofCancerPreventionandGenetics,IstitutoEuropeodiOncologia(IEO),Milan,Italy,91CogentechCancerGeneticTestLaboratory,Milan,Italy,92DepartmentofMedicalOncology,ErasmusUniversityMedicalCenter,Rotterdam,TheNetherlands,93DepartmentofClinicalGenetics,ErasmusUniversityMedicalCenter,Rotterdam,TheNetherlands,94DepartmentofGeneticsandPathology,PomeranianMedicalUniversity,Szczecin,Poland,95PostgraduateSchoolofMolecularMedicine,WarsawMedicalUniversity,Warsaw,Poland,96AnatomicalPathology,TheAlfredHospital,Melbourne,Australia,97Dr.
MargareteFischer-Bosch-InstituteofClinicalPharmacology,Stuttgart,Germany,98UniversityofTu¨bingen,Tu¨bingen,Germany,99InstituteforPreventionandOccupationalMedicineoftheGermanSocialAccidentInsurance,InstituteoftheRuhr-UniversityBochum(IPA),Bochum,Germany,100DepartmentofInternalMedicine,EvangelischeKlinikenBonngGmbH,JohanniterKrankenhaus,Bonn,Germany,101MolecularGeneticsofBreastCancer,GermanCancerResearchCenter(DKFZ),Heidelberg,Germany,102InstituteforOccupationalMedicineandMaritimeMedicine,UniversityMedicalCenterHamburg-Eppendorf,Hamburg,Germany,103InstituteofPathology,MedicalFacultyoftheUniversityofBonn,Bonn,Germany,104DivisionofClinicalEpidemiologyandAgingResearch,GermanCancerResearchCenter(DKFZ),Heidelberg,Germany,105GermanCancerConsortium(DKTK),Heidelberg,Germany,106SaarlandCancerRegistry,Saarbru¨cken,Germany,107DivisionofGeneticsandEpidemiologyandDivisionofBreastCancerResearch,TheInstituteofCancerResearch,Sutton,Surrey,UnitedKingdom,108CancerGenomicsLaboratory,CentreHospitalierUniversitairedeQuebecResearchCenterandLavalUniversity,Quebec,Canada,109DepartmentofMedicine,McGillUniversity,Montreal,Canada,110DivisionofClinicalEpidemiology,McGillUniversityHealthCentre,RoyalVictoriaHospital,Montreal,Quebec,Canada,111DepartementsdeSanteEnvironnementaleetSanteauTravailetdeMedecineSocialeetPreventive,UniversitedeMontreal,Montreal,Quebec,Canada,112LaboratoryofCancerGeneticsandTumorBiology,DepartmentofClinicalChemistryandBiocenterOulu,UniversityofOulu,NordLabOulu/OuluUniversityHospital,Oulu,Finland,113DepartmentofOncology,OuluUniversityHospital,UniversityofOulu,Oulu,Finland,114DepartmentofSurgery,OuluUniversityHospital,UniversityofOulu,Oulu,Finland,115SchoolofMedicine,InstituteofClinicalMedicine,Oncology,UniversityofEasternFinland,Kuopio,Finland,116CancerCenter,KuopioUniversityHospital,Kuopio,Finland,117SchoolofMedicine,InstituteofClinicalMedicine,PathologyandForensicMedicine,UniversityofEasternFinland,Kuopio,PLOSONE|www.
plosone.
org2November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRiskFinland,118ImagingCenter,DepartmentofClinicalPathology,KuopioUniversityHospital,Kuopio,Finland,119CancerCenterofEasternFinland,UniversityofEasternFinland,Kuopio,Finland,120DepartmentofGenetics,QIMRBerghoferMedicalResearchInstitute,Brisbane,AustraliaAbstractGeneticvariations,suchassinglenucleotidepolymorphisms(SNPs)inmicroRNAs(miRNA)orinthemiRNAbindingsitesmayaffectthemiRNAdependentgeneexpressionregulation,whichhasbeenimplicatedinvariouscancers,includingbreastcancer,andmayalterindividualsusceptibilitytocancer.
WeinvestigatedassociationsbetweenmiRNArelatedSNPsandbreastcancerrisk.
Firstweevaluated2,196SNPsinacase-controlstudycombiningninegenomewideassociationstudies(GWAS).
Second,wefurtherinvestigated42SNPswithsuggestiveevidenceforassociationusing41,785casesand41,880controlsfrom41studiesincludedintheBreastCancerAssociationConsortium(BCAC).
CombiningtheGWASandBCACdatawithinameta-analysis,weestimatedmaineffectsonbreastcancerriskaswellasrisksforestrogenreceptor(ER)andagedefinedsubgroups.
FivemiRNAbindingsiteSNPsassociatedsignificantlywithbreastcancerrisk:rs1045494(oddsratio(OR)0.
92;95%confidenceinterval(CI):0.
88–0.
96),rs1052532(OR0.
97;95%CI:0.
95–0.
99),rs10719(OR0.
97;95%CI:0.
94–0.
99),rs4687554(OR0.
97;95%CI:0.
95–0.
99,andrs3134615(OR1.
03;95%CI:1.
01–1.
05)locatedinthe39UTRofCASP8,HDDC3,DROSHA,MUSTN1,andMYCL1,respectively.
DROSHAbelongstomiRNAmachinerygenesandhasacentralroleininitialmiRNAprocessing.
Theremaininggenesareinvolvedindifferentmolecularfunctions,includingapoptosisandgeneexpressionregulation.
FurtherstudiesarewarrantedtoelucidatewhetherthemiRNAbindingsiteSNPsarethecausativevariantsfortheobservedriskeffects.
Citation:KhanS,GrecoD,MichailidouK,MilneRL,MuranenTA,etal.
(2014)MicroRNARelatedPolymorphismsandBreastCancerRisk.
PLoSONE9(11):e109973.
doi:10.
1371/journal.
pone.
0109973Editor:ZhongmingZhao,VanderbiltUniversityMedicalCenter,UnitedStatesofAmericaReceivedJune6,2014;AcceptedSeptember8,2014;PublishedNovember12,2014Thisisanopen-accessarticle,freeofallcopyright,andmaybefreelyreproduced,distributed,transmitted,modified,builtupon,orotherwiseusedbyanyoneforanylawfulpurpose.
TheworkismadeavailableundertheCreativeCommonsCC0publicdomaindedication.
DataAvailability:Theauthorsconfirmthat,forapprovedreasons,someaccessrestrictionsapplytothedataunderlyingthefindings.
DataareavailableviatheBreastCancerAssociationConsortium(BCAC)DataAccessCoordinationCommittee(DACC)(http://ccge.
medschl.
cam.
ac.
uk/consortia/bcac/).
Torequestthedata,readersmaycontactManjeetHumphreys(mkh39@medschl.
cam.
ac.
uk)orDouglasEaston(dfe20@medschl.
cam.
ac.
uk).
Funding:FundingfortheiCOGSinfrastructurecamefromtheEuropeanCommunity'sSeventhFrameworkProgrammeundergrantagreementnumber223175(HEALTH-F2-2009-223175)(COGS).
iCOGSwasalsopartlysupportedbytheCanadianInstitutesofHealthResearchforthe''CIHRTeaminFamilialRisksofBreastCancer''program(JS&DFE),andtheMinistryofEconomicDevelopment,InnovationandExportTradeofQuebec–grant#PSR-SIIRI-701(JS&DFE,P.
Hall).
HEBCSwasfinanciallysupportedbytheHelsinkiUniversityCentralHospitalResearchFund,AcademyofFinland(266528),theFinnishCancerSociety,TheNordicCancerUnionandtheSigridJuseliusFoundation.
ThepopulationalleleandgenotypefrequencieswereobtainedfromthedatasourcefundedbytheNordicCenterofExcellenceinDiseaseGeneticsbasedonsamplesregionallyselectedfromFinland,SwedenandDenmark.
TheUK2GWASwasfundedbyWellcomeTrustandCancerResearchUK.
ItincludedsamplescollectedthroughtheFBCSstudywhichisfundedbyCancerResearchUK[C8620/A8372].
TheWTCCCwasfundedbytheWellcomeTrust.
TheABCFSandOFBCRstudiesweresupportedbytheUnitedStatesNationalCancerInstitute,NationalInstitutesofHealth(NIH)underRFA-CA-06-503andthroughcooperativeagreementswithmembersoftheBreastCancerFamilyRegistry(BCFR)andPrincipalInvestigators,includingCancerCareOntario(U01CA69467),NorthernCaliforniaCancerCenter(U01CA69417),UniversityofMelbourne(U01CA69638).
SamplesfromtheNC-BCFRwereprocessedanddistributedbytheCoriellInstituteforMedicalResearch.
OFBCRwassupportedbytheCanadianInstitutesofHealthResearchforthe''CIHRTeaminFamilialRisksofBreastCancer''programandgrantUM1CA164920fromtheNationalCancerInstitute.
ThecontentofthismanuscriptdoesnotnecessarilyreflecttheviewsorpoliciesoftheNationalCancerInstituteoranyofthecollaboratingcentersintheBreastCancerFamilyRegistry(BCFR),nordoesmentionoftradenames,commercialproducts,ororganizationsimplyendorsementbytheUSGovernmentortheBCFR.
TheABCFSwasalsosupportedbytheNationalHealthandMedicalResearchCouncilofAustralia,theNewSouthWalesCancerCouncil,theVictorianHealthPromotionFoundation(Australia)andtheVictorianBreastCancerResearchConsortium.
JLHisaNationalHealthandMedicalResearchCouncil(NHMRC)AustraliaFellowandaVictorianBreastCancerResearchConsortiumGroupLeader.
MCSisaNHMRCSeniorResearchFellowandaVictorianBreastCancerResearchConsortiumGroupLeader.
JLHandMCSarebothgroupleadersoftheVictoriaBreastCancerResearchConsortium.
TheABCSstudywassupportedbytheDutchCancerSociety[grantsNKI2007-3839;20094363];BBMRI-NL,whichisaResearchInfrastructurefinancedbytheDutchgovernment(NWO184.
021.
007);andtheDutchNationalGenomicsInitiative.
TheBBCSisfundedbyCancerResearchUKandBreakthroughBreastCancerandacknowledgesNHSfundingtotheNIHRBiomedicalResearchCentre,andtheNationalCancerResearchNetwork(NCRN).
TheBBCSGWASreceivedfundingfromTheInstitutNationaldeCancer.
TheworkoftheBBCCwaspartlyfundedbyELAN-FondoftheUniversityHospitalofErlangen.
ES(BIGGS)issupportedbyNIHRComprehensiveBiomedicalResearchCentre,Guy's&St.
Thomas'NHSFoundationTrustinpartnershipwithKing'sCollegeLondon,UnitedKingdom.
ITissupportedbytheOxfordBiomedicalResearchCentre.
TheBSUCHstudywassupportedbytheDietmar-HoppFoundation,theHelmholtzSocietyandtheGermanCancerResearchCenter(DKFZ).
TheCECILEstudywasfundedbyFondationdeFrance[contractgrantnumber2004012618and2007005156],InstitutNationalduCancer(INCa)[2007-1/SPC2,2008-1-CP-4and2009-1-SHS/SP-04],LigueNationalecontreleCancer,AssociationpourlaRecherchecontreleCancer(ARC)[2008-1-CP-4];AgenceFrancaisedeSecuriteSanitairedel'EnvironnementetduTravail(AFSSET-ANSES)[ST-2005-003,EST2008/1/26,andVS-2009-21],LiguecontreleCancerGrandOuest.
TheCGPSwassupportedbytheChiefPhysicianJohanBoserupandLiseBoserupFund,theDanishMedicalResearchCouncilandHerlevHospital.
TheCNIO-BCSwassupportedbytheGenomeSpainFoundation,theRedTematicadeInvestigacionCooperativaenCancerandgrantsfromtheAsociacionEspanolaContraelCancerandtheFondodeInvestigacionSanitario(PI11/00923andPI081120).
WeacknowledgethesupportofAlvarezlvarez,DanielHerrero,PrimitivaMenendezandtheHumanGenotyping-CEGENUnit(CNIO).
TheHumanGenotyping-CEGENUnitissupportedbytheInstitutodeSaludCarlosIII.
TheCTSwassupportedbytheCaliforniaBreastCancerActof1993;NationalInstitutesofHealth(grantsR01CA77398andtheLonVSmithFoundation[LVS39420].
);andtheCaliforniaBreastCancerResearchFund(contract97-10500).
CollectionofcancerincidencedatausedinthisstudywassupportedbytheCaliforniaDepartmentofPublicHealthaspartofthestatewidecancerreportingprogrammandatedbyCaliforniaHealthandSafetyCodeSection103885.
DEMOKRITOSissupportedbyaHellenicCooperativeOncologyGroupresearchgrant(HRR_BG/04)andtheGreekGeneralSecretaryforResearchandTechnology(GSRT)Program,ResearchExcellenceII,fundedat75%bytheEuropeanUnion.
TheDFBBCSGWASwasfundedbyTheNetherlandsOrganisationforScientificResearch(NWO)aspartofaZonMw/VIDIgrantnumber91756341.
ThegenerationandmanagementofGWASgenotypedatafortheRotterdamStudyissupportedbytheNetherlandsOrganisationofScientificResearchNWOInvestments(nr.
175.
010.
2005.
011,911-03-012).
ThisstudyisfundedbytheResearchInstituteforDiseasesintheElderly(014-93-015;RIDE2),theNetherlandsGenomicsInitiative(NGI)/NetherlandsOrganisationforScientificResearch(NWO)projectnr.
050-060-810.
TheRotterdamStudyisfundedbyErasmusMedicalCenterandErasmusUniversity,Rotterdam,NetherlandsOrganizationfortheHealthResearchandDevelopment(ZonMw),theResearchInstituteforDiseasesintheElderly(RIDE),theMinistryofEducation,CultureandScience,theMinistryforHealth,WelfareandSports,theEuropeanCommission(DGXII),andtheMunicipalityofRotterdam.
TheESTHERstudywassupportdbyagrantfromtheBadenWu¨rttembergMinistryofScience,ResearchandArts.
AdditionalcaseswererecruitedinthecontextoftheVERDIstudy,whichwassupportedbyagrantfromtheGermanCancerAid(DeutscheKrebshilfe).
TheHMBCSwassupportedbyagrantfromtheFriendsofHannoverMedicalSchoolandbytheRudolfBartlingFoundation.
TheFinancialsupportforKARBACwasprovidedthroughtheregionalagreementonmedicaltrainingandclinicalresearch(ALF)betweenStockholmCountyCouncilandKarolinskaInstitutet,TheSwedishCancerSocietyandBertvonKantzowfoundation.
TheGC-HBOCwassupportedbyDeutscheKrebshilfe[107054],theDietmar-HoppFoundation,theHelmholtzsocietyandtheGermanCancerResearchCentre(DKFZ).
TheGC-HBOCGWASwasPLOSONE|www.
plosone.
org3November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRisksupportedbytheGermanCancerAid(grantno.
107352).
TheGENICAwasfundedbytheFederalMinistryofEducationandResearch(BMBF)Germanygrants01KW9975/5,01KW9976/8,01KW9977/0and01KW0114,theRobertBoschFoundation,Stuttgart,DeutschesKrebsforschungszentrum(DKFZ),Heidelberg,GermanSocialAccidentInsurance,InstituteoftheRuhrUniversityBochum(IPA),Germany,aswellastheDepartmentofInternalMedicine,EvangelischeKlinikenBonngGmbH,JohanniterKrankenhaus,Bonn,Germany.
TheKBCPwasfinanciallysupportedbythespecialGovernmentFunding(EVO)ofKuopioUniversityHospitalgrants,CancerFundofNorthSavo,theFinnishCancerOrganizations,andbythestrategicfundingoftheUniversityofEasternFinland.
kConFabissupportedbyagrantfromtheNationalBreastCancerFoundation,andpreviouslybytheNationalHealthandMedicalResearchCouncil(NHMRC),theQueenslandCancerFund,theCancerCouncilsofNewSouthWales,Victoria,TasmaniaandSouthAustralia,andtheCancerFoundationofWesternAustralia.
ThekConFabClinicalFollowUpStudywasfundedbytheNHMRC[145684,288704,454508].
FinancialsupportfortheAOCSwasprovidedbytheUnitedStatesArmyMedicalResearchandMaterielCommand[DAMD17-01-1-0729],theCancerCouncilofTasmaniaandCancerFoundationofWesternAustraliaandtheNHMRC[199600].
GCTissupportedbytheNHMRC.
LMBCissupportedbythe'StichtingtegenKanker'(232-2008and196-2010).
DietherLambrechtsissupportedbytheFWOandtheKULPFV/10/016-SymBioSysII.
TheMARIEstudywassupportedbytheDeutscheKrebshilfee.
V.
[70-2892-BRI],theHamburgCancerSociety,theGermanCancerResearchCenterandthegenotypeworkinpartbytheFederalMinistryofEducationandResearch(BMBF)Germany[01KH0402].
MBCSGissupportedbygrantsfromtheItalianAssociationforCancerResearch(AIRC)andbyfundsfromtheItaliancitizenswhoallocatedthe5/1000shareoftheirtaxpaymentinsupportoftheFondazioneIRCCSIstitutoNazionaleTumori,accordingtoItalianlaws(INT-Institutionalstrategicprojects''561000'').
TheMCBCSwassupportedbytheNIHgrants[CA122340,CA128978]andaSpecializedProgramofResearchExcellence(SPORE)inBreastCancer[CA116201],theBreastCancerResearchFoundationandagenerousgiftfromtheDavidF.
andMargaretT.
GrohneFamilyFoundationandtheTingTsungandWeiFongChaoFoundation.
MCCScohortrecruitmentwasfundedbyVicHealthandCancerCouncilVictoria.
TheMCCSwasfurthersupportedbyAustralianNHMRCgrants209057,251553and504711andbyinfrastructureprovidedbyCancerCouncilVictoria.
TheMECwassupportedbyNIHgrantsCA63464,CA54281,CA098758andCA132839.
FortheMTLGEBCSstudy,theinitialcase–controlstudywassupportedbytheCanadianBreastCancerResearchInitiative.
WorkwasalsosupportedbytheQuebecBreastCancerFoundation,theCanadianInstitutesofHealthResearchforthe''CIHRTeaminFamilialRisksofBreastCancer''program–grant#CRN-87521andtheMinistryofEconomicDevelopment,InnovationandExportTrade–grant#PSR-SIIRI-701.
TheNBCSwassupportedbygrantsfromtheNorwegianResearchcouncil,155218/V40,175240/S10toALBD,FUGE-NFR181600/V11toVNKandaSwizzBridgeAwardtoALBD.
TheOBCSwassupportedbyresearchgrantsfromtheFinnishCancerFoundation,theAcademyofFinland,theUniversityofOulu,andtheOuluUniversityHospital.
TheORIGOstudywassupportedbytheDutchCancerSociety(RUL1997-1505)andtheBiobankingandBiomolecularResourcesResearchInfrastructure(BBMRI-NLCP16).
TheOSUstudywasfundedbytheStefanieSpielmanfundandtheOSUComprehensiveCancerCenter.
ThePBCSwasfundedbyIntramuralResearchFundsoftheNationalCancerInstitute,DepartmentofHealthandHumanServices,USA.
ThepKARMAstudywassupportedbyMa¨ritandHansRausingsInitiativeAgainstBreastCancerandCancerRiskPredictionCenter,aLinneusCentre(contract70867902)financedbytheSwedishResearchCouncil.
TheRBCSwasfundedbytheDutchCancerSociety(DDHK2004-3124,DDHK2009-4318).
TheRPCIstudywassupportedbyRPCIDataBankandBioRepository(DBBR),aCancerCenterSupportGrantSharedResource(P30CA016056-32).
TheSASBACstudywassupportedbyfundingfromtheAgencyforScience,TechnologyandResearchofSingapore(A*STAR),theUSNationalInstituteofHealth(NIH)andtheSusanG.
KomenBreastCancerFoundation.
TheSBCSwassupportedbyYorkshireCancerResearchS295,S299,S305PA.
SEARCHisfundedbyaprogrammegrantfromCancerResearchUK[C490/A10124]andsupportedbythetheUKNationalInstituteforHealthResearchBiomedicalResearchCentreattheUniversityofCambridge.
AMDhasbeensupportedbyCancerResearchUKgrant[C8197/A10865]andbytheJosephMitchellFund.
SKKDKFZSissupportedbytheDKFZ.
TheSZBCSwassupportedbyGrantPBZ_KBN_122/P05/2004;KatarzynaJaworskaisafellowofInternationalPhDprogram,PostgraduateSchoolofMolecularMedicine,WarsawMedicalUniversity,supportedbythePolishFoundationofScience.
TheTNBCCwassupportedbytheNIHgrant[CA128978],theBreastCancerResearchFoundation,KomenFoundationfortheCure,theOhioStateUniversityComprehensiveCancerCenter,theStefanieSpielmanfundforBreastCancerResearchandagenerousgiftfromtheDavidF.
andMargaretT.
GrohneFamilyFoundationandtheTingTsungandWeiFongChaoFoundation.
PartoftheTNBCC(DEMOKRITOS)hasbeenco-financedbytheEuropeanUnion(EuropeanSocialFund–ESF)andGreeknationalfundsthroughtheOperationalProgram"EducationandLifelongLearning"oftheNationalStrategicReferenceFramework(NSRF)-ResearchFundingProgramoftheGeneralSecretariatforResearch&Technology:ARISTEIA.
TheUKBGSisfundedbyBreakthroughBreastCancerandtheInstituteofCancerResearch(ICR).
ICRacknowledgesNHSfundingtotheNIHRBiomedicalResearchCentre.
CGEMS,TheNurses'HealthStudiesaresupportedbyNIHgrantsCA65725,CA87969,CA49449,CA67262,CA50385and5UO1CA098233.
Thefundershadnoroleinstudydesign,datacollectionandanalysis,decisiontopublish,orpreparationofthemanuscript.
CompetingInterests:Theauthorshavedeclaredthatnocompetinginterestsexist.
*Email:heli.
nevanlinna@hus.
fi"MembershipoftheGENICANetwork,kConFabInvestigators,andAOCSisprovidedintheAcknowledgments.
IntroductionBreastcanceristhemostcommonwomen'scancerandisaleadingcauseofcancermortality[1].
Inheritedgeneticvariationhasbeenassociatedwiththeinitiation,developmentandprogressionofbreastcancer.
Studiesontwinshavesuggestedthathereditarypredisposingfactorsareinvolvedinuptoonethirdofallbreastcancers[2].
Manygeneticlocihavebeenassociatedwithbreastcancerriskandcollectivelyexplainapproximately35%ofthefamilialrisk[3,4].
Thelargestgeneticassociationstudyofbreastcancertodateidentified41novellowpenetrancesusceptibilityloci[4]byselectingnearly30,000SNPsfromameta-analysisofninegenome-wideassociation(GWA)studiesandgenotypingthemusing41,785casesand41,880controlsofEuropeanancestryfromstudiesintheBreastCancerAssociationConsortium(BCAC).
These41susceptibilitylociprobablyrepresentthetipoftheiceberg,andadditionalSNPsfromthecombinedGWASmightexplainasimilarfractionoffamilialrisktothatattributedtothealreadyidentifiedloci[4].
MaturemiRNAsare20–23nucleotide,single-strandedRNAmoleculesthatplayacrucialroleingeneexpressionregulationformanycellularprocessesincludingdifferentiationpotentialanddevelopmentpattern.
MiRNAsundergoastepwisematurationprocessinvolvinganarrayofmiRNAmachinerycomponents.
DroshaandDGCR8mediatethecleavageoflongprimarymiRNAtranscripts(pri-miRNAs)intoshorterpre-miRNAsinthenucleus[5,6].
Thepre-miRNAsarethentransportedtothecytoplasmwheretheyarefurthercleavedbyDicertoproducematuremiRNAs[7].
MiRNAsinteractbypairingwiththe39untranslatedregion(UTR),andalsowithinthecodingregionand59UTRofthecorrespondingmRNAsleadingtomRNAdestabilization,cleavageortranslationrepression.
MoreeffectivemRNAdestabilizationisachievedwhenmiRNAtargetsthe3'UTRratherthanothermRNAregions[8–10].
AnindividualmiRNAmayregulateapproximately100distinctmRNAs,andtogethermorethan1000humanmiRNAsarebelievedtomodulatemorethanhalfofthemRNAspeciesencodedinthegenome[11,12].
Additionally,mostmRNAspossessbindingsitesformiRNAs[13].
MiRNAsareinvolvedintumorigenesisinthattheycanbeeitheroncogenicwhentumorsuppressorgenesaretargeted,orgenomicguardians(tumoursuppressormiRNAs)whenoncogenesaretargeted[14].
Additionallyithasbeensuggestedthattheymaymodulatebothmetastasis[15]andchemotherapyresistance[16].
MiRNAshavealsobeenshowntohavealteredexpressionlevelsintumourscomparedtonormaltissueandbetweentumorsubtypesinbreastcanceramongothercarcinomatypes[17–19].
SNPsmayaffectmiRNAmachinerygenesormiRNAsactivity;howeverSNPscanalsocreate,abolishormodifymiRNAbindingsitesintheirbindingregions.
PolymorphismsinmiRNAbindingsiteshavebeenstudiedinregardtotheriskofseveralcancers[20],includingbreastcancer[21–23].
ThesestudieshavefoundevidenceforassociationofmiRNArelatedSNPsandcancerrisk,butthestudysamplesizeshavebeenrelativelysmall.
Inthisstudy,weinvestigateassociationsbetweenmiRNA-relatedpolymorphismsandbreastcancerriskbyusingameta-PLOSONE|www.
plosone.
org4November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRiskanalysisofnineGWASandsubsequentgenotypingoftophitsusing41,785casesand41,880controlsofEuropeanancestryfromtheBCAC.
Toourknowledge,thisisthusfarthelargestinvestigationofassociationsbetweenmiRNA-relatedpolymor-phismsandbreastcancersusceptibility.
MaterialsandMethodsSNPselectionandgenotypingSNPsinmatureorpre-miRNAs,ingenesofthemiRNAmachineryandin3'UTRregionsofproteincodinggeneswithapotentialeffectonmiRNAbindingweresystematicallysearchedfromEnsembl(hg18/build36)andPatroclesdatabases[24].
Additionally,taggingSNPsforsuchwithr2$0.
8werealsoidentifiedutilizingthepublicHapMapSNPdatabase.
Bythisinsilicoapproachweidentifiedaltogether147,801candidateSNPsand12,550taggingSNPs.
TheseSNPswerethenoverlayedwiththosefromthecombinedGWASfromtheBCAC[4]andaltogether2196SNPswerepresent(eithergenotypedorimputed)inthecombinedGWAS.
TheseSNPsweregenotypedwithIlluminaorAffymetrixarrays,asdescribedpreviously[25–32].
ThecombinedGWASdatawereimputedforallscansusingHapMapversion2CEUasareferenceinsimilarfashiontothatpresentedbyMichailidouandcolleagues[4]withtheexceptionthattheHapMapversion2release21wasusedatthetimetheoverlaywasperformed.
Analysisusinga1-degree-of-freedomtrendtestofthese2196SNPsinthecombinedGWASindicatedsomeevidenceofassociationwithbreastcancerriskfor44SNPs(p,0.
09).
Notably,thecombinedGWASincludedimputeddatageneratedusingHapMapversion2release21(basedonNCBIbuild35(dbSNPb125)),whereastheresultspresentedhereforthecombinedGWASarebasedonimputationusingHapMapversion2release22(basedonNCBIbuild36(dbSNPb126)).
Intherelease22,anumberofSNPswereexcludedduetomappinginconsistenciesinbuild35relativetobuild36.
Hence,theestimatesfromthecombinedGWASmayslightlydifferfromtheinitialassociationanalysis.
The44SNPs(including30candidateand14taggingSNP)weregenotypedonadditionalsamplesintheBCACusingthecustomIlluminaInfiniumarray(iCOGS)whichincludedatotalof211,155SNPsasdescribedpreviously.
ThedetaileddescriptionofqualitycontrolprocessforcombinedGWASandiCOGSgenotypingdatawaspresentedin[4].
Ofthe42SNPsthatpassedqualitycontrol[4],twowerelocatedinmiRNAgenes(onecandidateSNPlocatedinpre-miRNAhsa-miR-2110andonetagSNPtaggingamaturehsa-mir-548lvariant),andfourSNPswerelocatedinmiRNAmachinerygenes(SMAD5,SND1,CNOT4andDROSHA).
ThegenotypedDROSHASNPtagsthe39UTRmiRNAbindingsitevariantintheDROSHAgene.
Theremaining38candidateortagSNPswerelocatedin,ortaggedtoapredictedmiRNAbindingsiteinthe39UTRofproteincodinggenes.
All42SNPsaredescribedinTable1.
TheworkflowoftheSNPselectionindifferentstagesisillustratedinFigure1.
StudysampleThecombinedGWASincludedninebreastcancerstudiestotalling10,052casesand12,575controlsofEuropeanethnicbackground.
Detailsandstudy-specificsubjectnumbersarepresentedinTableS1.
SincetheGWASwerelimitedtopatientsofEuropeanethnicbackgroundwefurtherutilized41,785casesascertainedfortheirfirstprimary,invasivebreastcancerand41,880controlsofEuropeanancestryfrom41BCACstudiesgenotypedusingtheiCOGSarray(TableS2).
ForasubgroupanalysisofERnegativeandERpositivecases,aswellascasesagedlessthan50yearsatdiagnosis,weincludedallthecasesforwhichtherespectivedatawereavailable.
TheERsubgroupanalysiswasbasedon702ERnegativecasesand2,019ERpositivecasesfromfiveGWASstudiesand7,200ERnegativecasesfrom40BCACstudiesand26,302ERpositivecasesfrom34BCACstudies.
Theanalysisofcasesagedlessthan50yearsatdiagnosiswasbasedon3,470casesfromthreeGWASstudiesand9,483casesfrom35BCACstudies.
AllparticipatingstudiesconformtotheDeclarationofHelsinkiandwereapprovedbytherespectiveethicalreviewboardsandethicscommittees(TablesS1andS2),andallparticipantsinthesestudieshadprovidedwrittenconsentfortheresearch.
StatisticalmethodsWeusedlogisticregressiontoestimateper-allelelog-oddsratiosandstandarderrorsincludingthestudyasacovariate.
Wealsoincludedprincipalcomponentsascovariatesinordertocorrectforpotentialhiddenpopulationstructure.
IntheGWAS,fortwostudies(UK2andHEBCS)theestimateswereadjustedforthefirstthreeprincipalcomponentsandintheiCOGSanalysisweusedthefirstsixprincipalcomponentsandanadditionalcomponenttoreduceinflationfortheLMBCstudy,asdescribedpreviously[4].
SubgroupanalyseswerecarriedoutforERnegativeandpositivesubgroupsandforthegroupagedlessthan50yearsatdiagnosis.
Formeta-analysis,wecombinedtheestimatesfromthecombinedGWASandiCOGSwithafixedeffectsmodelusingtheinversevarianceweightedmethod.
Inthemeta-analysis,thesubjectsinvolvedinbothcombinedGWASandiCOGS(1880)wereonlytakenintoaccountonce.
InordertoadustforP-valuesagainstmultipletesting,weusedBenjaminiHochbergcorrection.
TheadjustedP-valuesareshowninTable2alongwiththenominalP-values.
InthetextwereportthenominalP-values.
ThestatisticalanalyseswereconductedusingtheR2.
14.
0statisticalcomputingenvironment(http://www.
r-project.
org/).
ResultsForthe42SNPswesuccessfullygenotyped,estimatesofassociationfromthecombinedGWASandfromiCOGSanalysisareshowninTableS3.
Twenty-oneSNPsshowedconsistentFigure1.
WorkflowofmiRNASNPselection.
doi:10.
1371/journal.
pone.
0109973.
g001PLOSONE|www.
plosone.
org5November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRiskTable1.
The42studiedSNPsinmiRNAs,miRNAmachinerygenesandmiRNAtargetgenes.
FunctionalSNP(TagSNP,R-squared)ChrPositionCodingGenemiRNASNPeffectaLocatedwithinmiRNArs1709140310115923895GAhsa-miR-2110rs13447640(rs1805360,r2=1)1193866677GAhsa-mir-548lLocatedinmiRNAbiogenesismachinerygenesrs37649415135497426ACSMAD5rs171516397127425052AGSND1rs174806167134773600CGCNOT4rs10719531437204GADROSHAhsa-miR-1298ACLocatedinmiRNAtargetgenesrs25503031654953111AGAMFRhsa-miR-577ACrs7513934152590776GACC2D1Bhsa-miR-384/hsa-miR-577CNCrs1128226721908194ACCDCA7Lhsa-miR-548gACrs37961333100000533GADCBLD2hsa-miR-624*ACrs74411290063806GADCNhsa-miR-135b*ACrs18034392137807312AGDYRK1Ahsa-miR-550ACrs37971527199858AGFAM189A1hsa-miR-570ACrs713062211128186721ACFLI1hsa-miR-138-2*ACrs10525321589275240AGHDDC3hsa-miR-1224-3p/hsa-miR-1260/hsa-miR-1280ACrs704012397160742AGKDM4Chsa-miR-154*/hsa-miR-487aACrs10622251049313232AGMAPK8hsa-miR-203ACrs417397116224740AGMEThsa-miR-576-5pACrs702681556253786AGMIER3hsa-miR-196a*ACrs3134615140134653CAMYCL1hsa-miR-1827ANCrs23046692238830402AGPER2hsa-miR-885-3pACrs134221715074900ACPMP22hsa-miR-29b-1*ACrs75623912201444411ACPPIL3hsa-miR-493*/hsa-miR-499-3pACrs7520333140862837AGRIMS3hsa-let-7d/hsa-let-7eCNCrs7396921853178524GAST8SIA3hsa-miR-96/hsa-miR-1271/hsa-miR-182ACrs10584504120200088GASYNPO2hsa-miR-183ACrs4351800117446395CASYT9hsa-miR-544ACrs124383241555366808AGTCF12hsa-miR-591ACrs128698701399415306GAZIC5hsa-miR-34a/hsa-miR-34c-5p/hsa-miR-449a/hsa-miR-449bACrs9990(rs1444418,r2=1)1064230476AGADOhsa-miR-512-5p/hsa-miR-510ACrs757537(rs4705870,r2=1)5132187033GAANKRD43hsa-miR-320a/hsa-miR-320b/hsa-miR-320c/hsa-miR-320dACrs3774729(rs2037119,r2=0.
943)363969919GAATXN7hsa-miR-1206ACPLOSONE|www.
plosone.
org6November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRiskassociationswithbreastcancerriskinthecombinedGWASandiniCOGSanalysis;resultsfromthemeta-analysisareshowninTable2.
ThemostsignificantlyassociatedSNP,rs702681(OR1.
06[95%CI1.
04–1.
08];P3.
9610210),islocatedinthe3'UTRofMIER3,closetotheknownbreastcancersusceptibilitygeneMAP3K1.
TheSNPrs702681islocatedatthesame5q11.
2locusasthepreviouslypublishedriskSNPrs889312[33](correlationr2=0.
3).
WhenthetwoSNPswereanalysedinthesamelogisticregressionmodel,theassociationwithrs889312,butnotthatwithrs702681remainednominallystatisticallysignificant,suggestingthatrs702681isunlikelytobethecausalSNPatthislocus.
ThefiveSNPswiththesignificantnovelassociationsfromthemeta-analysis(P#5.
0761023andadjustedP#3.
5561022aftercorrec-tionformultipletesting)werers1045494,(OR0.
92[95%CI0.
88–0.
96];P=5.
9061025),rs1052532,(OR0.
97[95%CI0.
95–0.
99];P=7.
7861024),rs10719,(OR0.
97[95%CI0.
94–0.
99];P=1.
3561023)rs4687554(OR0.
97[95%CI0.
95–0.
99];P=1.
7161023)andrs3134615(OR1.
03[95%CI1.
01–1.
05];P=5.
0761023)locatedin39UTRofCaspase-8(CASP8),HDDomainContaining3(HDDC3),DROSHA,Musculoskeletal,EmbryonicNuclearProtein1(MUSTN1)andV-MycMyelocy-tomatosisViralOncogeneHomolog1(MYCL1),respectively(Table2).
SNPrs1045494istaggingthehsa-miR-938bindingsiteSNPrs1045487(r2=1.
0)ofCASP8andtheSNPrs1052532inHDDC3ispredictedtoabolishthebindingsiteforhsa-miR-1224-3p.
TheSNPrs10719ispredictedtoabolishthehsa-miR-1298bindingsiteinthe39UTRofDROSHA.
SNPrs4687554tagsthehsa-miR-891bbindingsiteSNPrs6445538(r2=1.
0)ofMUSTN1andrs3134615islocatedatthebindingsiteofhsa-miR-1827ofMYCL1.
Therewasnoevidenceforheterogeneityintheper-alleleORforanySNP.
TheperstudyperalleleORsforthesefivemiRNAbindingsiteSNPsfromthecombinedGWASalongwithper-SNPheterogeneityvarianceP-valuesareshowninFigureS1andfromtheiCOGSinFigureS2.
NextweanalysedtheSNPsbyERstatus-definedsubtype,andforcasesagedlessthan50yearsatdiagnosis,forriskassociationsinthemeta-analysisofcombinedGWASandiCOGS(TablesS4,S5andS6).
Theseanalysesdidnotrevealanyadditionalsignificantresults.
Forrs1045494inCASP8,rs4687554inMUSTN1andrs3134615inMYCL1(OR1.
03[95%CI1.
01–1.
05];P=7.
7561024)amoresignificantassociationwithbreastcancerriskwasfoundfortheERpositivesubgroupthaninthemainanalysis,buttheresultfromthetestforheterogeneitybyERstatuswasnotsignificant(datanotshown).
Allassociationswereestimatedusinganadditiveinheritancemodel.
Dominantandrecessivemodelsdidnotimprovetheestimates(datanotshown).
DiscussionWeinvestigatedassociationsbetweengeneticvariationinmiRNAs,inthegenesofthemiRNAmachineryandinthemiRNAbindingsitesandtheriskofbreastcancer.
WeidentifiedseveralSNPsthatarepredictedtoabolishanmiRNAbindingsiteandthataresignificantlyassociatedwithbreastcancerrisk.
PreviousstudiesinvestigatingmiRNArelatedSNPs,especiallyinmiRNAbindingsiteshaveincludedpredefinedsetsofgenes.
Nicolosoandcolleaguesinvestigated38previouslyidentifiedbreastcancerriskSNPsandfoundtwotomodifymiRNAbindingsitesinTGFB1andXRCC1invitro[23].
Neitherofthesewereincludedinourdataset.
Liangandcolleaguesinvestigated134potentialmiRNAbindingsitesincancer-relatedgenesandfoundsixmiRNAbindingsiteSNPsthatwereassociatedwithovariancancerrisk[34].
Table1.
Cont.
FunctionalSNP(TagSNP,R-squared)ChrPositionCodingGenemiRNASNPeffectars1045487(rs1045494,r2=1)2201860026AGCASP8hsa-miR-938ACrs7288826(rs8140217,r21)2237547947GACBX6hsa-miR-1207-5pACrs17569034(rs17512204,r2=0.
835)2118449301GACCDC93hsa-miR-1178ACrs3205281(rs7674744,r2=1)478874296GACNOT6Lhsa-miR-643/hsa-miR-297ACrs13005(rs9473,r2=0.
964)1013727177GAFRMD4Ahsa-miR-548mACrs3809831(rs3809828,r2=1)177187575GAKCTD11hsa-miR-892bACrs6445538(rs4687554,r2=1)352839175AGMUSTN1hsa-miR-891bACrs7818(rs9371201,r2=0.
875)6150186694GAPCMT1hsa-miR-595ACrs9844202(rs7635553,r2=1)3168646064GASERPINI2hsa-miR-1272ACrs2271565(rs7086917,r2=1)1049867441ACWDFY4hsa-miR-657/hsa-miR-214/hsa-miR-15a/hsa-miR-16/hsa-miR-15b/hsa-miR-195/hsa-miR-424/hsa-miR-497ACTagSNPsusedintheanalysisarepresentedintheparenthesisalongwiththeRsquredvaluerelativetothefunctionalSNP.
aAccordingtoPatroclesprediction;AC=abolishesconservedbindingsite,ANC=abolishesnon-conservedbindingsite,CNC=createsnon-conservedbindingsite(Targetsitesareconsideredconservediftheyaresharedbyatleastoneprimate,onerodentandonenonprimate/nonrodentmammal[24]).
doi:10.
1371/journal.
pone.
0109973.
t001PLOSONE|www.
plosone.
org7November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRiskTable2.
AssociationsofSNPsintheGWASandiCOGSseparatelyandcombinedGWAS+iCOGSandbreastcancerrisk.
SNPChrPositioncoding1GWASOR(95%CI)2GWASP3iCOGSOR(95%CI)2iCOGSP3CombinedGWAS+iCOGSOR(95%CI)2CombinedGWAS+iCOGSP3(BHcorrectedP)4Geners702681556253786AG1.
07(1.
02–1.
11)3.
92610231.
06(1.
04–1.
09)2.
76610281.
06(1.
04–1.
08)3.
88610210(1.
6361028)MIER3rs10454942201860026AG0.
90(0.
81–1.
00)4.
74610220.
92(0.
88–0.
96)4.
47610240.
92(0.
88–0.
96)5.
9461025(1.
2561023)CASP8rs10525321589275240AG0.
94(0.
90–0.
98)7.
94610230.
97(0.
95–0.
99)1.
47610220.
97(0.
95–0.
99)7.
7861024(1.
0961022)HDDC3rs10719531437204GA0.
92(0.
88–0.
97)8.
79610240.
98(0.
95–1.
00)5.
32610220.
97(0.
94–0.
99)1.
3561023(1.
4261022)DROSHArs4687554352839175AG0.
94(0.
90–0.
99)1.
23610220.
97(0.
95–1.
00)2.
39610220.
97(0.
95–0.
99)1.
7161023(1.
4461022)MUSTN1rs3134615140134653CA1.
04(0.
99–1.
09)9.
97610221.
03(1.
00–1.
05)2.
09610221.
03(1.
01–1.
05)5.
0761023(3.
5561022)MYCL1rs76355533168646064GA0.
89(0.
83–0.
95)9.
73610240.
98(0.
95–1.
01)1.
98610211.
00(0.
97–1.
04)9.
2461023(5.
5461022)SERPINI2rs37961333100000533GA1.
18(1.
08–1.
29)4.
18610241.
01(0.
97–1.
06)5.
74610211.
04(1.
00–1.
09)3.
9361022(1.
4561021)DCBLD2rs4351800117446395CA1.
04(1.
00–1.
08)4.
48610221.
01(0.
99–1.
03)1.
98610211.
02(1.
00–1.
04)4.
1561022(1.
4561021)SYT9rs175122042118449301GA1.
06(0.
98–1.
14)1.
20610211.
03(0.
99–1.
06)1.
63610211.
03(1.
00–1.
07)5.
2261022(1.
5761021)CCDC93rs3809828177187575GA1.
17(1.
06–1.
28)1.
97610231.
01(0.
97–1.
05)5.
22610210.
99(0.
95–1.
03)7.
9361022(2.
2261021)KCTD11rs74411290063806GA1.
11(1.
03–1.
20)8.
70610231.
01(0.
97–1.
05)5.
98610211.
03(0.
99–1.
06)1.
0461021(2.
5761021)DCNrs70869171049867441AC0.
96(0.
93–1.
00)6.
35610220.
99(0.
97–1.
01)4.
38610210.
99(0.
97–1.
00)1.
2961021(3.
0161021)WDFY4rs704012397160742AG1.
11(0.
99–1.
23)7.
59610221.
02(0.
97–1.
07)5.
14610211.
00(0.
95–1.
04)1.
7961021(3.
7461021)KDM4Crs7674744478874296GA0.
94(0.
89–0.
99)2.
83610220.
99(0.
97–1.
02)6.
91610211.
01(0.
98–1.
03)1.
8161021(3.
7461021)CNOT6Lrs124383241555366808AG0.
87(0.
79–0.
97)1.
01610221.
00(0.
94–1.
05)8.
69610211.
02(0.
98–1.
07)1.
8761021(3.
7461021)TCF12rs171516397127425052AG0.
96(0.
92–1.
01)1.
09610210.
99(0.
97–1.
02)5.
66610211.
00(0.
98–1.
02)2.
1961021(4.
1861021)SND1rs174806167134773600CG0.
87(0.
72–1.
04)1.
27610210.
99(0.
93–1.
04)6.
39610210.
98(0.
92–1.
03)3.
7061021(5.
9861021)CNOT4rs7513934152590776GA1.
04(1.
00–1.
08)7.
98610221.
00(0.
98–1.
02)9.
99610211.
01(0.
99–1.
02)4.
3761021(6.
3461021)CC2D1Brs23046692238830402AG0.
96(0.
91–1.
02)1.
86610211.
00(0.
97–1.
02)8.
17610210.
99(0.
97–1.
02)4.
3861021(6.
3461021)PER2rs10584504120200088GA0.
96(0.
91–1.
01)1.
33610211.
00(0.
97–1.
02)9.
28610211.
01(0.
98–1.
03)4.
5961021(6.
4361021)SYNPO2TheSNPswithconsistentoddsratiosincombinedGWASandiCOGSanalysisareshown.
(Resultsforall42SNPsarepresentedinTableS3.
)1Build36position.
2Peralleleoddsratiofortheminorallelerelativetothemajorallele.
31dfp-trend.
41dfp-trendadjustedagainstmultipletestingbyBenjamini–Hochbergcorrectionmethod.
doi:10.
1371/journal.
pone.
0109973.
t002PLOSONE|www.
plosone.
org8November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRiskInthemeta-analysisofcombinedGWASandiCOGSformaineffects,forfourofthefivemostsignificantmiRNAbindingsiteSNPs,theminorallelewasassociatedwithadecreasedbreastcancerrisk.
TheminoralleleofSNPrs3134615in39UTRofMYCL1wasassociatedwithanincreasedbreastcancerrisk.
AllthefivemostsignificantmiRNAbindingsiteSNPslocatein39UTRandhavebeenpredictedtoabolishthemiRNAbindingsite.
ThedefectinmiRNA-mediatedregulationwouldbeexpectedtoleadtoanincreaseinthetranslationofthecorrespondingencodedprotein.
Thefivegenes,whoseregulationmaybeaffectedbythemiRNA-associatedSNPs,includethepre-apoptoticgeneCASP8,HDDC3,miRNAbiogenesismasterregulatorDROSHA,MYC-familymemberMYCL1andMUSTN1.
CASP8isinvolvedinapoptosisinbreastcancercells[35],andmanystudieshavereportedpolymorphismsinthisgenetobeassociatedwithrisksforseveralcancers[36,37]includingbreastcancer[38,39],indicatingtheimportanceofCASP8intumordevelopment.
SNPrs1045494studiedhereislocatedclosetothecodingregionSNPrs1045485thathasbeenpreviouslyshowntohaveastrongerprotectiveeffect[38,40,41].
Interestingly,MichalidouandcolleaguesreportedthisSNPashavingonlyweakevidenceforanassociation(P0.
0013incombinedGWASandiCOGS)[4],butthesetwoSNPs(rs1045485andrs1045494)arenotcorrelated(r2=0.
001inCaucasianpopulation).
Neitherisrs1045494correlatedwiththemorestronglyassociatedrs1830298SNP,identifiedthroughfine-mappingoftheregion(r2=0.
02)[42].
Rs1045494tagsSNPrs1045487(r2=1.
0)whichispredictedtoabolishthehsa-miR-938bindingsiteandthusmayaffectCASP8expression.
ThereisverylittlereportedevidenceontheinvolvementofHDDC3orthehsa-miR-1224-3pincancer,indicatinganovelassociationwithrisk.
HDDC3hasbeensuggestedtobeinvolvedinthestarvationresponse[43].
TheHDDC3geneisexpressedathigherlevelsbyseveraldifferenttumortypes,includingbreasttumors,thanbynormaltissue[44].
DROSHAisamiRNAmasterregulator.
ItisamemberoftheRNaseIIIenzymefamily,belongstothemiRNAbiogenesispathwayandisthecorenucleasethatprocessespri-miRNAsintopre-miRNAsinthenucleus[5,6].
TheSNPrs10719inthe39UTRofDROSHAispredictedtoabolishthehsa-miR-1298bindingsite.
Hsa-miR-1298ispredictedtotargetDROSHAbythePatroclespredictionaswellasbyTargetScan[45]andPITA[46]predictionalgorithms.
RecentlyasmallKoreanstudyreportedanotherSNPrs644236,taggingtheSNPrs10719(r2=0.
955inCEUpopulationandr2=0.
876inAsianpopulation(combinedCHBandJPT))tobeassociatedwithelevatedbreastcancerrisk[47].
WhentakingintoaccounttheoppositemajorandminorsallelesintheAsianandEuropeanpopulationsforSNPsrs644236andrs10719,thisresultisinconcordancewithourresultswhereboththecombinedGWASaswellastheiCOGSanalysisconsistentlyindicatedanassociationoftheminoralleleofSNPrs10719withreducedbreastcancerrisk.
WealsofoundtheminoralleleofSNPrs3134615inthe39UTRofMYCL1tobeassociatedwithanincreasedrisk.
MYCL1(L-MYC)belongstothesamefamilyoftranscriptionfactorsastheknownproto-oncogeneMYC(C-MYC)andtheyshareahighdegreeofstructuralsimilarity[48].
TheMYCL1genehaspreviouslybeenreportedtobeamplifiedandoverexpressedinovariancancer[49].
Acase-controlstudybyXiongandcolleaguesreportedSNPrs3134615tobesignificantlyassociatedwithincreasedriskofsmallcelllungcancer[50].
SNPrs3134615waspredictedbyPatroclestoabolishthehsa-miR-1827bindingsite.
ThishasalsobeensuggestedbyfunctionalstudieswhereMYCL1wasfoundasthetargetofhsa-miR-1827andtheSNPrs3134615wasalsofoundtoincreaseMYCL1expression[50].
TheevidencefromfunctionalstudiesisconsistentwithourfindingthatSNPrs3134615mightincreasebreastcancerrisk.
MUSTN1hasbeenshowntobeinvolvedinthedevelopmentandregenerationofthemusculoskeletalsystem[51].
ThusfarnoevidenceofassociationbetweenMUSTN1andbreastcancerhasbeenreported,buttheMUSTN1geneisexpressedinthemammaryglands[52].
SinceonlyasmallfractionofmiRNAbindingsiteshasbeenexperimentallyvalidated,weselectedSNPsthathadbeencomputationallypredictedtoaffectmiRNAbindingsites.
ForouroriginalSNPselectionweusedthePatroclesdatabasethatcontainspredictedmiRNAbindingsitesandalsocompilesperturbationpredictionofSNPeffects.
Thereareamultitudeofpredictionprogramsandtheirperformancehasbeenevaluated[53].
Witkosandcolleaguesfindtargetpredictionalgorithmsthatutilizeorthologoussequencealignment,likePatrocles,tobethemostreliable.
Thefollowupofthe42miRNArelatedSNPsidentifiedfivesignificantassociationswithbreastcancerrisk.
Althoughtheindividualriskeffectsweresubtle,consideringthatwecouldonlyinvestigateasmallproportionofourinitialinsilicodatasetofmiRNArelatedSNPs(over140,000SNPs)thismaysuggestthatgeneticpolymorphismsaffectingthemiRNAregulationcouldhaveaconsiderablecombinedeffectonbreastcancerrisk.
Itshouldbenotedthat,untilfinemappingstudiesarecarriedoutfortheseloci,itisnotclearwhetherthesemiRNA-relatedSNPsarethevariantsresponsiblefortheobservedassociations.
ThiscomprehensiveanalysisofmiRNArelatedpolymorphismsusingalargetwostagestudyofwomenwithEuropeanancestryprovidesevidenceformiRNArelatedSNPsbeingpotentialmodulatorsofbreastcancerrisk.
SupportingInformationFigureS1ForestplotsforthefivemostsignificantmiRNAbindingsiteSNPsfromthecombinedGWAS.
Squaresindicatetheestimatedper-alleleORfortheminoralleleinEuropeans.
Thehorizontallinesindicate95%confidencelimits.
Theverticalbluedashedlinesindicateclippingoftheconfidenceintervalsforpresentationpurpose.
Theareaofthesquareisinverselyproportionaltothevarianceoftheestimate.
Thediamondindicatestheestimatedper-alleleORfromthecombinedanalysis.
(PDF)FigureS2ForestplotsforthefivemostsignificantmiRNAbindingsiteSNPsfromtheiCOGS.
Squaresindicatetheestimatedper-alleleORfortheminoralleleinEuropeans.
Thehorizontallinesindicate95%confidencelimits.
Theverticalbluedashedlinesindicateclippingoftheconfidenceintervalsforpresentationpurpose.
Theareaofthesquareisinverselyproportionaltothevarianceoftheestimate.
Thediamondindicatestheestimatedper-alleleORfromthecombinedanalysis.
(PDF)TableS1AdescriptionofeachGWASstudy,numberofsubjectsandgenotypingplatformusedincombinedGWAS.
(DOC)TableS2AdescriptionofeachBCACstudywithsubjectsofEuropeanorigininiCOGS.
(DOC)TableS3Frequenciesandeffectsizesofthe42SNPsinthemainanalysis;combinedGWASandiCOGS.
(DOC)PLOSONE|www.
plosone.
org9November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRiskTableS4ResultsforSNPsintheGWASandiCOGSseparatelyandcombinedGWAS+iCOGSanalysisforERnegativesubgroup.
(DOC)TableS5ResultsforSNPsintheGWASandiCOGSseparatelyandcombinedGWAS+iCOGSanalysisforERpositivesubgroup.
(DOC)TableS6ResultsforSNPsintheGWASandiCOGSseparatelyandcombinedGWAS+iCOGSanalysisforcaseslessthan50yearsatdiagnosis.
(DOC)AcknowledgmentsWethankalltheindividualswhotookpartinthesestudiesandalltheresearchers,studystaff,cliniciansandotherhealthcareproviders,techniciansandadministrativestaffwhohaveenabledthisworktobecarriedout.
TheHEBCSthanksDr.
KarlvonSmittenandRNIrjaErkkila¨fortheirhelpwiththeHEBCSdataandsamples.
TheABCFSthanksMaggieAngelakos,JudiMaskiellandGillianDite.
TheOFBCRthanksTeresaSelander,NayanaWeerasooriyaandGordGlendon.
TheABCSwouldliketoacknowledgeEllenvanderSchootforDNAofcontrols.
TheBBCCthanksSilkeLandrith,SonjaOeser,MatthiasRu¨bner.
TheBBCSthanksEileenWilliams,ElaineRyder-MillsandKaraSargus.
TheBIGGSthanksNiallMcInerney,GabrielleColleran,AndrewRowanandAngelaJones.
TheBSUCHthanksPeterBugertandtheMedicalFaculty,Mannheim.
TheCGPSthanksthestaffandparticipantsoftheCopenhagenGeneralPopulationStudy,andDortheUldallAndersen,MariaBirnaArnadottir,AnneBank,DortheKjeldgardHansenforexcellenttechnicalassistance.
TheCNIO-BCSacknowledgethesupportofNuriaAlvarez,DanielHerrero,PrimitivaMenendezandtheHumanGenotyping-CEGENUnit(CNIO).
TheDFBBCSthanksMargreetAusems,ChristivanAsperen,SennoVerhoef,andRogiervanOldenburgforprovidingsamplesfromtheirClinicalGeneticcenters.
WealsothankPascalArp,MilaJhamai,MarijnVerkerk,LizbethHerreraandMarjoleinPetersfortheirhelpincreatingtheGWASdatabase,andKarolEstradaandMaksimV.
Struchalinfortheirsupportincreationandanalysisofimputeddata.
Theauthorsaregratefultothestudyparticipants,thestafffromtheRotterdamStudyandtheparticipatinggeneralpractitionersandpharmacists.
TheESTHERthanksHartwigZiegler,SonjaWolfandVolkerHermann,KatjaButterbach.
TheGC-HBOCwouldliketothankthefollowingpersonsforprovidingadditionalinformationandsamples:Prof.
Dr.
NorbertArnold,Dr.
SabinePreissler-Adams,Dr.
MonikaMareeva-Varon,Dr.
DieterNiederacher,Prof.
Dr.
BrigitteSchlegelberger,Dr.
ClemensMu¨l,HeideHellebrand,andStefanieEngert.
TheHMBCSthanksPeterHillemanns,HansChristiansenandJohannH.
Karstens.
TheKBCPthanksEijaMyo¨ha¨nenandHelenaKemila¨inen.
kConFab/AOCSwishtothankHeatherThorne,EvelineNiedermayr,allthekConFabresearchnursesandstaff,theheadsandstaffoftheFamilyCancerClinics,andtheClinicalFollowUpStudyfortheircontributionstothisresource,andthemanyfamilieswhocontributetokConFab.
TheLMBCthanksGilianPeuteman,DominiekSmeets,ThomasVanBrusselandKathleenCorthouts.
TheMARIEwouldliketothankAlinaVrieling,KatharinaBuck,UrsulaEilber,MuhabbetCelik,andSabineBehrens.
TheMBCSGthanksSiranoushManoukian,BernardPeisselandDanielaZaffaronioftheFondazioneIRCCSIstitutoNazionaledeiTumori(INT);BernardoBonanni,IreneFeroceandAngelaManiscalcooftheIstitutoEuropeodiOncologia(IEO)andthepersonneloftheCogentechCancerGeneticTestLaboratory.
TheMTLGEBCSgratefullyacknowledgetheassistanceofLesleyRichardsonandMarie-ClaireGouletinconductingthestudy.
WewouldliketothankMartineTranchant(CancerGenomicsLaboratory,CHUdeQuebecResearchCenter),Marie-FranceValois,AnnieTurgeonandLeaHeguy(McGillUniversityHealthCenter,RoyalVictoriaHospital;McGillUniversity)forDNAextraction,samplemanagementandskillfultechnicalassistance.
J.
S.
isChairholderoftheCanadaResearchChairinOncogenetics.
TheOBCSthanksMeeriOtsukkaandKariMononen.
TheORIGOthanksE.
Krol-Warmerdam,andJ.
Blomforpatientaccrual,administeringquestionnaires,andmanagingclinicalinformation.
TheLUMCsurvivaldatawereretrievedfromtheLeidenhospital-basedcancerregistrysystem(ONCDOC)withthehelpofDr.
J.
Molenaar.
TheOSUthanksRobertPilarksiandCharlesShapiro,whowereinstrumentalintheformationoftheOSUBreastCancerTissueBank.
WethanktheHumanGeneticsSampleBankforprocessingofsamples.
OSUColumbusareacontrolspecimenswereprovidedbytheOhioStateUniversity'sHumanGeneticsSampleBank.
ThePBCSthanksMarkSherman,NeonilaSzeszenia-Dabrowska,BeataPeplonska,WitoldZatonski,PeiChaoandMichaelStagner.
TheRBCSthanksPetraBos,JannetBlom,EllenCrepin,AnjaNieuwlaat,AnnetteHeemskerkandtheErasmusMCFamilyCancerClinic.
TheSBCSthanksSueHigham,IanBrock,SabapathyBalasu-bramanian,HelenCrampandDanConnley.
TheSEARCHthankstheSEARCHandEPIC-Norfolkteams.
TheiCOGSstudywouldnothavebeenpossiblewithoutthecontributionsofthefollowing:QinWang(BCAC),AndrewBerchuck(OCAC),RosalindA.
Eeles,AliAminAlOlama,ZsofiaKote-Jarai,SaraBenlloch(PRACTICAL),AntonisAntoniou,LesleyMcGuffogandKenOffit(CIMBA),AndrewLee,andEdDicks,CraigLuccariniandthestaffoftheCentreforGeneticEpidemiologyLaboratory,AnnaGonzalez-NeiraandthestaffoftheCNIOgenotypingunit,DanielC.
Tessier,FrancoisBacot,DanielVincent,SylvieLaBoissie`reandFredericRobidouxandthestaffoftheMcGillUniversityandGenomeQuebecInnovationCentre,andthestaffoftheCopenhagenDNAlaboratory,andJulieM.
Cunningham,SharonA.
Windebank,ChristopherA.
Hilker,JeffreyMeyerandthestaffofMayoClinicGenotypingCoreFacility.
ConsortiamembersGENICANetwork.
HiltrudBrauch,Wing-YeeLo,ChristinaJustenhoven:Dr.
MargareteFischer-Bosch-InstituteofClinicalPharma-cology,Stuttgart,andUniversityofTu¨bingen,Germany.
Yon-DschunKo,ChristianBaisch:DepartmentofInternalMedicine,EvangelischeKlinikenBonngGmbH,JohanniterKrankenhaus,Bonn,Germany.
Hans-PeterFischer:InstituteofPathology,UniversityofBonn,Bonn,Germany.
UteHamann:MolecularGeneticsofBreastCancer,DeutschesKrebs-forschungszentrum(DKFZ),Heidelberg,Germany.
ThomasBru¨ning,BeatePesch,SylviaRabstein,AnneLotz:InstituteoftheRuhrUniversityBochum(IPA),Bochum,Germany.
VolkerHarth:InstituteforOccupa-tionalMedicineandMaritimeMedicine,UniversityMedicalCenterHamburg-Eppendorf,Germany.
kConFabInvestigators.
Seehttp://www.
kconfab.
org/Organisation/Members.
aspxAOCS.
Seehttp://www.
aocstudy.
org/org_coll.
aspAuthorContributionsConceivedanddesignedtheexperiments:HNDGGCTACRLMDFESKKMJCCADMSMGCPH.
Performedtheexperiments:SKDGKMRLMDFE.
Analyzedthedata:SKDGKMRLMHNDFE.
Contributedreagents/materials/analysistools:SKHNDGKMGCTACRLMPDPPUHMKSA.
MeindlRWTHCBK.
AaltonenGGGDFEPAFMJHILAH.
BrauchQWEJSH.
BrennerAKDMSGFLTAMK.
Aittoma¨kiJ.
LiuPHAIKHJ.
LiKCJCCRHARPSDFJOFJPIdSSNJLGZAJLHHTM.
BuiEMDFSMCSCAJ.
StoneHMHMAARBvdLA.
MannermaaRKSBMMPLCTNRSJCDJHSSCMWRRABLJVVFBHMGSABEMWBSEBBGNSFNHFPMZJIAPJ.
BenitezCAHBEHFSLLMAMDMSRLJ.
BrownFJCXWCVJEODLMMRPMRCPGTTPLPC.
MulotFMA.
SchneeweissC.
SohnBBITMJKNMJAKSTAMMNVBNNATDHACHDMEMGCJFJ.
LissowskaLBPDRAEMTC.
SeynaeveCJvAVNKSSAETCBADYALSMPRPPM.
BarilePMJWMMJMCA.
JagerA.
JakubowskaJ.
LubinskiKJBKDC.
McLeanTBYDKVAC.
StegmaierA.
SwerdlowAANOMJJ.
SimardMDKPAJVMGVKMKBJDVMKJMHkConFabInvestigatorsAustralianOvarianCancerStudyGroupTheGENICANetwork.
Wrotethepaper:SKHNRLMAC.
Providedcriticalreviewofthemanuscript:SKHNDGKMGCTACRLMPDPPUHMKSA.
MeindlRWTHCBK.
AaltonenGGGDFEPAFMJHILAH.
BrauchQWEJSH.
BrennerAKDMSGFLTAMK.
Aittoma¨kiJ.
LiuPHAIKHJ.
LiKCJCCRHARPSDFJOFJPIdSSNJLGZAJLHHTM.
BuiEMDFSMCSCAJ.
StoneHMHMAARBvdLA.
MannermaaRKSBMMPLCTNRSJCDJHSSCMWRRABLJVVFBHMGSABEMWBSEBBGNSFNHFPMZJIAPJ.
BenitezCAHBEHFSLLMAMDMSRLJ.
BrownFJCXWCVJEODLMMRPMRCPGTTPLPC.
MulotFMA.
SchneeweissC.
SohnBBITMJKNMJAKSTAMMNVBNNATDHACHDMEMGCJFJ.
LissowskaLBPDRAEMTC.
SeynaeveCJvAVNKSSAETCBADYALPLOSONE|www.
plosone.
org10November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRiskSMPRPPM.
BarilePMJWMMJMCA.
JagerA.
JakubowskaJ.
LubinskiKJBKDC.
McLeanTBYDKVAC.
StegmaierA.
SwerdlowAANOMJJ.
SimardMDKPAJVMGVKMKBJDVMKJMHkConFabInvestigatorsAustralianOvarianCancerStudyGroupTheGENICANetwork.
Approvedthefinalversionofthemanuscript:SKHNDGKMGCTACRLMPDPPUHMKSA.
MeindlRWTHCBK.
AaltonenGGGDFEPAFMJHILAH.
BrauchQWEJSH.
BrennerAKDMSGFLTAMK.
Aittoma¨kiJ.
LiuPHAIKHJ.
LiKCJCCRHARPSDFJOFJPIdSSNJLGZAJLHHTM.
BuiEMDFSMCSCAJ.
StoneHMHMAARBvdLA.
MannermaaRKSBMMPLCTNRSJCDJHSSCMWRRABLJVVFBHMGSABEMWBSEBBGNSFNHFPMZJIAPJ.
BenitezCAHBEHFSLLMAMDMSRLJ.
BrownFJCXWCVJEODLMMRPMRCPGTTPLPC.
MulotFMA.
SchneeweissC.
SohnBBITMJKNMJAKSTAMMNVBNNATDHACHDMEMGCJFJ.
LissowskaLBPDRAEMTC.
SeynaeveCJvAVNKSSAETCBADYALSMPRPPM.
BarilePMJWMMJMCA.
JagerA.
JakubowskaJ.
LubinskiKJBKDC.
McLeanTBYDKVAC.
StegmaierA.
SwerdlowAANOMJJ.
SimardMDKPAJVMGVKMKBJDVMKJMHkConFabInvestigatorsAustralianOvarianCancerStudyGroupTheGENICANetwork.
Administrativetechnicalormaterialsupport:MKBJDMSRL.
References1.
JemalA,BrayF,CenterMM,FerlayJ,WardE,etal.
(2011)Globalcancerstatistics.
CACancerJClin61:69–90.
2.
LichtensteinP,HolmNV,VerkasaloPK,IliadouA,KaprioJ,etal.
(2000)Environmentalandheritablefactorsinthecausationofcancer—analysesofcohortsoftwinsfromSweden,Denmark,andFinland.
NEnglJMed343:78–85.
3.
GhoussainiM,FletcherO,MichailidouK,TurnbullC,SchmidtMK,etal.
(2012)Genome-wideassociationanalysisidentifiesthreenewbreastcancersusceptibilityloci.
NatGenet44:312–318.
4.
MichailidouK,HallP,Gonzalez-NeiraA,GhoussainiM,DennisJ,etal.
(2013)Large-scalegenotypingidentifies41newlociassociatedwithbreastcancerrisk.
NatGenet45:353–361,361e351–352.
5.
DenliAM,TopsBB,PlasterkRH,KettingRF,HannonGJ(2004)ProcessingofprimarymicroRNAsbytheMicroprocessorcomplex.
Nature432:231–235.
6.
LeeY,AhnC,HanJ,ChoiH,KimJ,etal.
(2003)ThenuclearRNaseIIIDroshainitiatesmicroRNAprocessing.
Nature425:415–419.
7.
HutvagnerG,McLachlanJ,PasquinelliAE,BalintE,TuschlT,etal.
(2001)AcellularfunctionfortheRNA-interferenceenzymeDicerinthematurationofthelet-7smalltemporalRNA.
Science293:834–838.
8.
SosioM,KloostermanH,BianchiA,deVreugdP,DijkhuizenL,etal.
(2004)OrganizationoftheteicoplaningeneclusterinActinoplanesteichomyceticus.
Microbiology150:95–102.
9.
FilipowiczW,BhattacharyyaSN,SonenbergN(2008)Mechanismsofpost-transcriptionalregulationbymicroRNAs:aretheanswersinsightNatRevGenet9:102–114.
10.
ShuklaGC,SinghJ,BarikS(2011)MicroRNAs:Processing,Maturation,TargetRecognitionandRegulatoryFunctions.
MolCellPharmacol3:83–92.
11.
KrolJ,LoedigeI,FilipowiczW(2010)ThewidespreadregulationofmicroRNAbiogenesis,functionanddecay.
NatRevGenet11:597–610.
12.
ZhongX,CoukosG,ZhangL(2012)miRNAsinhumancancer.
MethodsMolBiol822:295–306.
13.
FriedmanRC,FarhKK,BurgeCB,BartelDP(2009)MostmammalianmRNAsareconservedtargetsofmicroRNAs.
GenomeRes19:92–105.
14.
FaraziTA,HoellJI,MorozovP,TuschlT(2013)MicroRNAsinHumanCancer.
AdvExpMedBiol774:1–20.
15.
WangX,ChenX,WangR,XiaoP,XuZ,etal.
(2013)microRNA-200cmodulatestheepithelial-to-mesenchymaltransitioninhumanrenalcellcarcinomametastasis.
OncolRep30:643–650.
16.
LiangZ,WuH,XiaJ,LiY,ZhangY,etal.
(2010)InvolvementofmiR-326inchemotherapyresistanceofbreastcancerthroughmodulatingexpressionofmultidrugresistance-associatedprotein1.
BiochemPharmacol79:817–824.
17.
VoliniaS,CroceCM(2013)PrognosticmicroRNA/mRNAsignaturefromtheintegratedanalysisofpatientswithinvasivebreastcancer.
ProcNatlAcadSciUSA110:7413–7417.
18.
GuoL,ZhaoY,YangS,CaiM,WuQ,etal.
(2012)Genome-widescreenforaberrantlyexpressedmiRNAsrevealsmiRNAprofilesignatureinbreastcancer.
MolBiolRep.
19.
(2012)Comprehensivemolecularportraitsofhumanbreasttumours.
Nature490:61–70.
20.
LandiD,GemignaniF,NaccaratiA,PardiniB,VodickaP,etal.
(2008)Polymorphismswithinmicro-RNA-bindingsitesandriskofsporadiccolorectalcancer.
Carcinogenesis29:579–584.
21.
SongF,ZhengH,LiuB,WeiS,DaiH,etal.
(2009)AnmiR-502-bindingsitesingle-nucleotidepolymorphisminthe3'-untranslatedregionoftheSET8geneisassociatedwithearlyageofbreastcanceronset.
ClinCancerRes15:6292–6300.
22.
KontorovichT,LevyA,KorostishevskyM,NirU,FriedmanE(2010)SinglenucleotidepolymorphismsinmiRNAbindingsitesandmiRNAgenesasbreast/ovariancancerriskmodifiersinJewishhigh-riskwomen.
IntJCancer127:589–597.
23.
NicolosoMS,SunH,SpizzoR,KimH,WickramasingheP,etal.
(2010)Single-nucleotidepolymorphismsinsidemicroRNAtargetsitesinfluencetumorsusceptibility.
CancerRes70:2789–2798.
24.
HiardS,CharlierC,CoppietersW,GeorgesM,BaurainD(2010)Patrocles:adatabaseofpolymorphicmiRNA-mediatedgeneregulationinvertebrates.
NucleicAcidsRes38:D640–651.
25.
DiteGS,JenkinsMA,SoutheyMC,HockingJS,GilesGG,etal.
(2003)Familialrisks,early-onsetbreastcancer,andBRCA1andBRCA2germlinemutations.
JNatlCancerInst95:448–457.
26.
FletcherO,JohnsonN,PallesC,dosSantosSilvaI,McCormackV,etal.
(2006)InconsistentassociationbetweentheSTK15F31Igeneticpolymorphismandbreastcancerrisk.
JNatlCancerInst98:1014–1018.
27.
HunterDJ,KraftP,JacobsKB,CoxDG,YeagerM,etal.
(2007)Agenome-wideassociationstudyidentifiesallelesinFGFR2associatedwithriskofsporadicpostmenopausalbreastcancer.
NatGenet39:870–874.
28.
FrankB,HemminkiK,WappenschmidtB,MeindlA,KlaesR,etal.
(2006)AssociationoftheCASP10V410IvariantwithreducedfamilialbreastcancerriskandinteractionwiththeCASP8D302Hvariant.
Carcinogenesis27:606–609.
29.
Flesch-JanysD,SlangerT,MutschelknaussE,KroppS,ObiN,etal.
(2008)Riskofdifferenthistologicaltypesofpostmenopausalbreastcancerbytypeandregimenofmenopausalhormonetherapy.
IntJCancer123:933–941.
30.
LiJ,HumphreysK,HeikkinenT,AittomakiK,BlomqvistC,etal.
(2011)Acombinedanalysisofgenome-wideassociationstudiesinbreastcancer.
BreastCancerResTreat126:717–727.
31.
LeuM,HumphreysK,SurakkaI,RehnbergE,MuiluJ,etal.
(2010)NordicDB:aNordicpoolandportalforgenome-widecontroldata.
EurJHumGenet18:1322–1326.
32.
TurnbullC,AhmedS,MorrisonJ,PernetD,RenwickA,etal.
(2010)Genome-wideassociationstudyidentifiesfivenewbreastcancersusceptibilityloci.
NatGenet42:504–507.
33.
EastonDF,PooleyKA,DunningAM,PharoahPD,ThompsonD,etal.
(2007)Genome-wideassociationstudyidentifiesnovelbreastcancersusceptibilityloci.
Nature447:1087–1093.
34.
LiangD,MeyerL,ChangDW,LinJ,PuX,etal.
(2010)GeneticvariantsinMicroRNAbiosynthesispathwaysandbindingsitesmodifyovariancancerrisk,survival,andtreatmentresponse.
CancerRes70:9765–9776.
35.
Ruiz-RuizC,Munoz-PinedoC,Lopez-RivasA(2000)Interferon-gammatreatmentelevatescaspase-8expressionandsensitizeshumanbreasttumorcellstoadeathreceptor-inducedmitochondria-operatedapoptoticprogram.
CancerRes60:5673–5680.
36.
BarrettJH,IlesMM,HarlandM,TaylorJC,AitkenJF,etal.
(2011)Genome-wideassociationstudyidentifiesthreenewmelanomasusceptibilityloci.
NatGenet43:1108–1113.
37.
deMartinoM,HaitelA,SchatzlG,KlinglerHC,KlatteT(2013)TheCASP8-6526NInsertion/DeletionPromoterPolymorphismIsAssociatedwithRenalCellCarcinomaRiskandMetastasis.
JUrol.
38.
CoxA,DunningAM,Garcia-ClosasM,BalasubramanianS,ReedMW,etal.
(2007)AcommoncodingvariantinCASP8isassociatedwithbreastcancerrisk.
NatGenet39:352–358.
39.
PengS,LuB,RuanW,ZhuY,ShengH,etal.
(2011)Geneticpolymorphismsandbreastcancerrisk:evidencefrommeta-analyses,pooledanalyses,andgenome-wideassociationstudies.
BreastCancerResTreat127:309–324.
40.
MacPhersonG,HealeyCS,TeareMD,BalasubramanianSP,ReedMW,etal.
(2004)AssociationofacommonvariantoftheCASP8genewithreducedriskofbreastcancer.
JNatlCancerInst96:1866–1869.
41.
FrankB,BermejoJL,HemminkiK,KlaesR,BugertP,etal.
(2005)Re:AssociationofacommonvariantoftheCASP8genewithreducedriskofbreastcancer.
JNatlCancerInst97:1012;authorreply1012–1013.
42.
LinWY,CampNJ,GhoussainiM,BeesleyJ,MichailidouK,etal.
(2014)IdentificationandcharacterizationofnovelassociationsintheCASP8/ALS2CR12regiononchromosome2withbreastcancerrisk.
HumMolGenet.
43.
SunD,LeeG,LeeJH,KimHY,RheeHW,etal.
(2010)AmetazoanorthologofSpoThydrolyzesppGppandfunctionsinstarvationresponses.
NatStructMolBiol17:1188–1194.
44.
KilpinenS,AutioR,OjalaK,IljinK,BucherE,etal.
(2008)Systematicbioinformaticanalysisofexpressionlevelsof17,330humangenesacross9,783samplesfrom175typesofhealthyandpathologicaltissues.
GenomeBiol9:R139.
45.
LewisBP,BurgeCB,BartelDP(2005)Conservedseedpairing,oftenflankedbyadenosines,indicatesthatthousandsofhumangenesaremicroRNAtargets.
Cell120:15–20.
46.
KerteszM,IovinoN,UnnerstallU,GaulU,SegalE(2007)TheroleofsiteaccessibilityinmicroRNAtargetrecognition.
NatGenet39:1278–1284.
47.
SungH,LeeKM,ChoiJY,HanS,LeeJY,etal.
(2011)CommongeneticpolymorphismsofmicroRNAbiogenesispathwaygenesandriskofbreastcancer:acase-controlstudyinKorea.
BreastCancerResTreat130:939–951.
PLOSONE|www.
plosone.
org11November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRisk48.
BirrerMJ,SegalS,DeGreveJS,KayeF,SausvilleEA,etal.
(1988)L-myccooperateswithrastotransformprimaryratembryofibroblasts.
MolCellBiol8:2668–2673.
49.
WuR,LinL,BeerDG,EllensonLH,LambBJ,etal.
(2003)AmplificationandoverexpressionoftheL-MYCproto-oncogeneinovariancarcinomas.
AmJPathol162:1603–1610.
50.
XiongF,WuC,ChangJ,YuD,XuB,etal.
(2011)GeneticvariationinanmiRNA-1827bindingsiteinMYCL1alterssusceptibilitytosmall-celllungcancer.
CancerRes71:5175–5181.
51.
LombardoF,KomatsuD,HadjiargyrouM(2004)MolecularcloningandcharacterizationofMustang,anovelnuclearproteinexpressedduringskeletaldevelopmentandregeneration.
FASEBJ18:52–61.
52.
KapusheskyM,AdamusiakT,BurdettT,CulhaneA,FarneA,etal.
(2012)GeneExpressionAtlasupdate—avalue-addeddatabaseofmicroarrayandsequencing-basedfunctionalgenomicsexperiments.
NucleicAcidsRes40:D1077–1081.
53.
WitkosTM,KoscianskaE,KrzyzosiakWJ(2011)PracticalAspectsofmicroRNATargetPrediction.
CurrMolMed11:93–109.
PLOSONE|www.
plosone.
org12November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRisk
Milne4,5,TaruA.
Muranen1,TuomasHeikkinen1,KirsimariAaltonen1,6,7,JoeDennis3,ManjeetK.
Bolla3,JianjunLiu8,PerHall9,AstridIrwanto8,KeithHumphreys9,JingmeiLi8,KamilaCzene9,JennyChang-Claude10,RebeccaHein10,11,AnjaRudolph10,PetraSeibold10,DieterFlesch-Janys12,OliviaFletcher13,JulianPeto14,IsabeldosSantosSilva14,NicholaJohnson13,LornaGibson14,ZoeAitken14,JohnL.
Hopper15,HelenTsimiklis16,MinhBui15,EnesMakalic15,DanielF.
Schmidt15,MelissaC.
Southey16,CarmelApicella15,JenniferStone15,QuintenWaisfisz17,HanneMeijers-Heijboer17,MurielA.
Adank17,RobB.
vanderLuijt18,AlfonsMeindl19,RitaK.
Schmutzler20,21,22,23,BertramMu¨ller-Myhsok24,PeterLichtner25,ClareTurnbull26,NazneenRahman26,StephenJ.
Chanock27,DavidJ.
Hunter28,29,AngelaCox30,SimonS.
Cross31,MalcolmW.
R.
Reed30,MarjankaK.
Schmidt32,AnnegienBroeks32,LauraJ.
Van'tVeer32,FransB.
Hogervorst32,PeterA.
Fasching33,34,MichaelG.
Schrauder33,ArifB.
Ekici35,MatthiasW.
Beckmann33,StigE.
Bojesen36,37,BrgeG.
Nordestgaard36,37,SuneF.
Nielsen36,37,HenrikFlyger38,JavierBenitez39,40,PilarM.
Zamora41,JoseI.
A.
Perez42,ChristopherA.
Haiman43,BrianE.
Henderson43,FredrickSchumacher43,LoicLeMarchand44,PaulD.
P.
Pharoah3,45,AlisonM.
Dunning45,MitulShah45,RobertLuben46,JudithBrown3,FergusJ.
Couch47,XianshuWang47,CelineVachon48,JanetE.
Olson48,DietherLambrechts49,50,MatthieuMoisse49,50,RobertParidaens51,Marie-RoseChristiaens51,PascalGuenel52,53,There`seTruong52,53,PierreLaurent-Puig54,ClaireMulot54,FrederickMarme55,56,BarbaraBurwinkel55,57,AndreasSchneeweiss55,56,ChristofSohn55,ElinorJ.
Sawyer58,IanTomlinson59,MichaelJ.
Kerin60,NicolaMiller60,IreneL.
Andrulis61,62,JuliaA.
Knight63,64,SandrineTchatchou61,AnnaMarieMulligan65,66,ThiloDo¨rk67,NataliaV.
Bogdanova68,NataliaN.
Antonenkova69,HodaAnton-Culver70,HatefDarabi9,MikaelEriksson9,MontserratGarcia-Closas71,72,JonineFigueroa27,JolantaLissowska73,LouiseBrinton27,PeterDevilee74,RobertA.
E.
M.
Tollenaar75,CarolineSeynaeve76,ChristiJ.
vanAsperen77,VesselaN.
Kristensen78,79,80,kConFabInvestigators81",AustralianOvarianCancerStudyGroup81,82",SusanSlager48,AmandaE.
Toland83,ChristineB.
Ambrosone84,DrakoulisYannoukakos85,AnnikaLindblom86,SaraMargolin87,PaoloRadice88,PaoloPeterlongo89,MonicaBarile90,PaoloMariani89,91,MaartjeJ.
Hooning92,JohnW.
M.
Martens92,J.
MargrietCollee93,AgnesJager92,AnnaJakubowska94,JanLubinski94,KatarzynaJaworska-Bieniek94,95,KatarzynaDurda94,GrahamG.
Giles4,5,CatrionaMcLean96,HiltrudBrauch97,98,ThomasBru¨ning99,Yon-DschunKo100,TheGENICANetwork97,98,99,100,101,102,103",HermannBrenner104,105,AidaKarinaDieffenbach104,105,VolkerArndt104,ChristaStegmaier106,AnthonySwerdlow107,AlanAshworth13,NickOrr13,MichaelJones71,JacquesSimard108,MarkS.
Goldberg109,110,FranceLabre`che111,MartineDumont108,RobertWinqvist112,KatriPylka¨s112,ArjaJukkola-Vuorinen113,MerviGrip114,VesaKataja115,116,Veli-MattiKosma117,118,119,JaanaM.
Hartikainen117,118,119,ArtoMannermaa117,118,119,UteHamann101,GeorgiaChenevix-Trench120,CarlBlomqvist7,KristiinaAittoma¨ki6,DouglasF.
Easton3,45,HeliNevanlinna1*1DepartmentofObstetricsandGynecology,UniversityofHelsinkiandHelsinkiUniversityCentralHospital,Helsinki,Finland,2FinnishInstituteofOccupationalHealth,Helsinki,Finland,3CentreforCancerGeneticEpidemiology,DepartmentofPublicHealthandPrimaryCare,UniversityofCambridge,Cambridge,UnitedKingdom,4CancerEpidemiologyCentre,CancerCouncilVictoria,Melbourne,Australia,5CentreforEpidemiologyandBiostatistics,MelbourneSchoolofPopulationandGlobalHealth,TheUniversityofMelbourne,Melbourne,Australia,6DepartmentofClinicalGenetics,UniversityofHelsinkiandHelsinkiUniversityCentralHospital,Helsinki,Finland,7DepartmentofOncology,UniversityofHelsinkiandHelsinkiUniversityCentralHospital,Helsinki,Finland,8HumanGeneticsDivision,GenomeInstituteofSingapore,Singapore,Singapore,9DepartmentofMedicalEpidemiologyandBiostatistics,KarolinskaInstitutet,Stockholm,Sweden,10DivisionofCancerEpidemiology,GermanCancerResearchCenter(DKFZ),Heidelberg,Germany,11PMVResearchGroupattheDepartmentofChildandAdolescentPsychiatryandPsychotherapy,UniversityofCologne,Cologne,Germany,12DepartmentofCancerEpidemiology/ClinicalCancerRegistryandInstituteforMedicalBiometricsandEpidemiology,UniversityClinicHamburg-Eppendorf,Hamburg,Germany,13BreakthroughBreastCancerResearchCentre,TheInstituteofCancerResearch,London,UnitedKingdom,14DepartmentofNon-CommunicableDiseaseEpidemiologyDepartment,LondonSchoolofHygieneandTropicalMedicine,London,UnitedKingdom,15CentreforPLOSONE|www.
plosone.
org1November2014|Volume9|Issue11|e109973EpidemiologyandBiostatistics,MelbourneSchoolofPopulationandGlobalHealth,TheUniversityofMelbourne,Melbourne,Australia,16DepartmentofPathology,TheUniversityofMelbourne,Melbourne,Australia,17DepartmentofClinicalGenetics,VUUniversityMedicalCenter,Amsterdam,TheNetherlands,18DepartmentofMedicalGenetics,UniversityMedicalCenterUtrecht,Utrecht,TheNetherlands,19DivisionofGynaecologyandObstetrics,TechnischeUniversita¨tMu¨nchen,Munich,Germany,20DivisionofMolecularGyneco-Oncology,DepartmentofGynaecologyandObstetrics,UniversityHospitalofCologne,Cologne,Germany,21CenterofFamilialBreastandOvarianCancer,UniversityHospitalofCologne,Cologne,Germany,22CenterforIntegratedOncology(CIO),UniversityHospitalofCologne,Cologne,Germany,23CenterforMolecularMedicineCologne(CMMC),UniversityofCologne,Cologne,Germany,24MaxPlanckInstituteofPsychiatry,Munich,Germany,25InstituteofHumanGenetics,HelmholtzZentrumMu¨nchen,GermanResearchCenterforEnvironmentalHealth,Neuherberg,Germany,26SectionofCancerGenetics,InstituteofCancerResearch,Sutton,UnitedKingdom,27DivisionofCancerEpidemiologyandGenetics,NationalCancerInstitute,Rockville,Maryland,UnitedStatesofAmerica,28PrograminMolecularandGeneticEpidemiology,HarvardSchoolofPublicHealth,Boston,Massachusetts,UnitedStatesofAmerica,29DepartmentofEpidemiology,HarvardSchoolofPublicHealth,Boston,Massachusetts,UnitedStatesofAmerica,30CRUK/YCRSheffieldCancerResearchCentre,DepartmentofOncology,UniversityofSheffield,Sheffield,UnitedKingdom,31AcademicUnitofPathology,DepartmentofNeuroscience,UniversityofSheffield,Sheffield,UnitedKingdom,32NetherlandsCancerInstitute,AntonivanLeeuwenhoekhospital,Amsterdam,TheNetherlands,33UniversityBreastCenterFranconia,DepartmentofGynecologyandObstetrics,UniversityHospitalErlangen,Friedrich-AlexanderUniversityErlangen-Nuremberg,ComprehensiveCancerCancerErlangen-EMN,Erlangen,Germany,34DavidGeffenSchoolofMedicine,DepartmentofMedicineDivisionofHematologyandOncology,UniversityofCaliforniaLosAngeles,California,UnitedStatesofAmerica,35InstituteofHumanGenetics,UniversityHospitalErlangen,Friedrich-AlexanderUniversityErlangen-Nuremberg,ComprehensiveCancerCenterErlangen-EMN,Erlangen,Germany,36CopenhagenGeneralPopulationStudy,HerlevHospital,CopenhagenUniversityHospital,Copenhagen,Denmark,37DepartmentofClinicalBiochemistry,HerlevHospital,CopenhagenUniversityHospital,Copenhagen,Denmark,38DepartmentofBreastSurgery,HerlevHospital,CopenhagenUniversityHospital,Copenhagen,Denmark,39HumanGeneticsGroup,HumanCancerGeneticsProgram,SpanishNationalCancerResearchCentre(CNIO),Madrid,Spain,40CentrodeInvestigacionenReddeEnfermedadesRaras(CIBERER),Valencia,Spain,41ServiciodeOncologaMedica,HospitalUniversitarioLaPaz,Madrid,Spain,42ServiciodeCirugaGeneralyEspecialidades,HospitalMonteNaranco,Oviedo,Spain,43DepartmentofPreventiveMedicine,KeckSchoolofMedicine,UniversityofSouthernCalifornia,LosAngeles,California,UnitedStatesofAmerica,44EpidemiologyProgram,CancerResearchCenter,UniversityofHawaii,Honolulu,Hawaii,UnitedStatesofAmerica,45CentreforCancerGeneticEpidemiology,DepartmentofOncology,UniversityofCambridge,Cambridge,UnitedKingdom,46ClinicalGerontology,DepartmentofPublicHealthandPrimaryCare,UniversityofCambridge,Cambridge,UnitedKingdom,47DepartmentofLaboratoryMedicineandPathology,MayoClinic,Rochester,Minnesota,UnitedStatesofAmerica,48DepartmentofHealthSciencesResearch,MayoClinic,Rochester,Minnesota,UnitedStatesofAmerica,49VesaliusResearchCenter(VRC),VIB,Leuven,Belgium,50LaboratoryforTranslationalGenetics,DepartmentofOncology,UniversityofLeuven,Leuven,Belgium,51OncologyDepartment,UniversityHospitalGasthuisberg,Leuven,Belgium,52Inserm(NationalInstituteofHealthandMedicalResearch),CESP(CenterforResearchinEpidemiologyandPopulationHealth),U1018,EnvironmentalEpidemiologyofCancer,Villejuif,France,53UniversityParis-Sud,UMRS1018,Villejuif,France,54UniversiteParisSorbonneCite,UMR-S775Inserm,Paris,France,55DepartmentofObstetricsandGynecology,UniversityofHeidelberg,Heidelberg,Germany,56NationalCenterforTumorDiseases,UniversityofHeidelberg,Heidelberg,Germany,57MolecularEpidemiologyGroup,GermanCancerResearchCenter(DKFZ),Heidelberg,Germany,58ResearchOncology,DivisionofCancerStudies,King'sCollegeLondon,Guy'sHospital,London,UnitedKingdom,59WellcomeTrustCentreforHumanGeneticsandOxfordBiomedicalResearchCentre,UniversityofOxford,Oxford,UnitedKingdom,60ClinicalScienceInstitute,UniversityHospitalGalway,Galway,Ireland,61Lunenfeld-TanenbaumResearchInstituteofMountSinaiHospital,Toronto,Ontario,Canada,62DepartmentofMolecularGenetics,UniversityofToronto,Toronto,Ontario,Canada,63ProssermanCentreforHealthResearch,Lunenfeld-TanenbaumResearchInstitute,MountSinaiHospital,Toronto,Ontario,Canada,64DivisionofEpidemiology,DallaLanaSchoolofPublicHealth,UniversityofToronto,Toronto,Ontario,Canada,65DepartmentofLaboratoryMedicineandPathobiology,UniversityofToronto,Toronto,Ontario,Canada,66DepartmentofLaboratoryMedicine,andtheKeenanResearchCentreoftheLiKaShingKnowledgeInstitute,StMichael'sHospital,Toronto,Ontario,Canada,67DepartmentofObstetricsandGynaecology,HannoverMedicalSchool,Hannover,Germany,68DepartmentofRadiationOncology,HannoverMedicalSchool,Hannover,Germany,69N.
N.
AlexandrovResearchInstituteofOncologyandMedicalRadiology,Minsk,Belarus,70DepartmentofEpidemiology,UniversityofCaliforniaIrvine,Irvine,California,UnitedStatesofAmerica,71DivisionofGeneticsandEpidemiology,InstituteofCancerResearch,Sutton,UnitedKingdom,72BreakthroughBreastCancerResearchCentre,DivisionofBreastCancerResearch,TheInstituteofCancerResearch,London,UnitedKingdom,73DepartmentofCancerEpidemiologyandPrevention,M.
Sklodowska-CurieMemorialCancerCenterandInstituteofOncology,Warsaw,Poland,74DepartmentofHumanGenetics&DepartmentofPathology,LeidenUniversityMedicalCenter,Leiden,Netherlands,75DepartmentofSurgicalOncology,LeidenUniversityMedicalCenter,Leiden,Netherlands,76FamilyCancerClinic,DepartmentofMedicalOncology,ErasmusMC-DanieldenHoedCancerCenter,Rotterdam,Netherlands,77DepartmentofClinicalGenetics,LeidenUniversityMedicalCenter,Leiden,Netherlands,78InstituteofClinicalMedicine,FacultyofMedicine,UniversityofOslo,Oslo,Norway,79DepartmentofClinicalMolecularBiology(EpiGen),UniversityofOslo,Oslo,Norway,80DepartmentofGenetics,InstituteforCancerResearch,OsloUniversityHospital,Radiumhospitalet,Oslo,Norway,81PeterMacCallumCancerCenter,Melbourne,Australia,82QIMRBerghoferMedicalResearchInstitute,Brisbane,Australia,83DepartmentofMolecularVirology,ImmunologyandMedicalGenetics,ComprehensiveCancerCenter,TheOhioStateUniversity,Columbus,Ohio,UnitedStatesofAmerica,84RoswellParkCancerInstitute,Buffalo,NewYork,UnitedStatesofAmerica,85MolecularDiagnosticsLaboratory,IRRP,NationalCentreforScientificResearch"Demokritos",AghiaParaskeviAttikis,Athens,Greece,86DepartmentofMolecularMedicineandSurgery,KarolinskaInstitutet,Stockholm,Sweden,87DepartmentofOncology-Pathology,KarolinskaInstitutet,Stockholm,Sweden,88UnitofMolecularBasesofGeneticRiskandGeneticTesting,DepartmentofPreventiveandPredictiveMedicine,FondazioneIRCCSIstitutoNazionaledeiTumori(INT),Milan,Italy,89IFOM,FondazioneIstitutoFIRCdiOncologiaMolecolare,Milan,Italy,90DivisionofCancerPreventionandGenetics,IstitutoEuropeodiOncologia(IEO),Milan,Italy,91CogentechCancerGeneticTestLaboratory,Milan,Italy,92DepartmentofMedicalOncology,ErasmusUniversityMedicalCenter,Rotterdam,TheNetherlands,93DepartmentofClinicalGenetics,ErasmusUniversityMedicalCenter,Rotterdam,TheNetherlands,94DepartmentofGeneticsandPathology,PomeranianMedicalUniversity,Szczecin,Poland,95PostgraduateSchoolofMolecularMedicine,WarsawMedicalUniversity,Warsaw,Poland,96AnatomicalPathology,TheAlfredHospital,Melbourne,Australia,97Dr.
MargareteFischer-Bosch-InstituteofClinicalPharmacology,Stuttgart,Germany,98UniversityofTu¨bingen,Tu¨bingen,Germany,99InstituteforPreventionandOccupationalMedicineoftheGermanSocialAccidentInsurance,InstituteoftheRuhr-UniversityBochum(IPA),Bochum,Germany,100DepartmentofInternalMedicine,EvangelischeKlinikenBonngGmbH,JohanniterKrankenhaus,Bonn,Germany,101MolecularGeneticsofBreastCancer,GermanCancerResearchCenter(DKFZ),Heidelberg,Germany,102InstituteforOccupationalMedicineandMaritimeMedicine,UniversityMedicalCenterHamburg-Eppendorf,Hamburg,Germany,103InstituteofPathology,MedicalFacultyoftheUniversityofBonn,Bonn,Germany,104DivisionofClinicalEpidemiologyandAgingResearch,GermanCancerResearchCenter(DKFZ),Heidelberg,Germany,105GermanCancerConsortium(DKTK),Heidelberg,Germany,106SaarlandCancerRegistry,Saarbru¨cken,Germany,107DivisionofGeneticsandEpidemiologyandDivisionofBreastCancerResearch,TheInstituteofCancerResearch,Sutton,Surrey,UnitedKingdom,108CancerGenomicsLaboratory,CentreHospitalierUniversitairedeQuebecResearchCenterandLavalUniversity,Quebec,Canada,109DepartmentofMedicine,McGillUniversity,Montreal,Canada,110DivisionofClinicalEpidemiology,McGillUniversityHealthCentre,RoyalVictoriaHospital,Montreal,Quebec,Canada,111DepartementsdeSanteEnvironnementaleetSanteauTravailetdeMedecineSocialeetPreventive,UniversitedeMontreal,Montreal,Quebec,Canada,112LaboratoryofCancerGeneticsandTumorBiology,DepartmentofClinicalChemistryandBiocenterOulu,UniversityofOulu,NordLabOulu/OuluUniversityHospital,Oulu,Finland,113DepartmentofOncology,OuluUniversityHospital,UniversityofOulu,Oulu,Finland,114DepartmentofSurgery,OuluUniversityHospital,UniversityofOulu,Oulu,Finland,115SchoolofMedicine,InstituteofClinicalMedicine,Oncology,UniversityofEasternFinland,Kuopio,Finland,116CancerCenter,KuopioUniversityHospital,Kuopio,Finland,117SchoolofMedicine,InstituteofClinicalMedicine,PathologyandForensicMedicine,UniversityofEasternFinland,Kuopio,PLOSONE|www.
plosone.
org2November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRiskFinland,118ImagingCenter,DepartmentofClinicalPathology,KuopioUniversityHospital,Kuopio,Finland,119CancerCenterofEasternFinland,UniversityofEasternFinland,Kuopio,Finland,120DepartmentofGenetics,QIMRBerghoferMedicalResearchInstitute,Brisbane,AustraliaAbstractGeneticvariations,suchassinglenucleotidepolymorphisms(SNPs)inmicroRNAs(miRNA)orinthemiRNAbindingsitesmayaffectthemiRNAdependentgeneexpressionregulation,whichhasbeenimplicatedinvariouscancers,includingbreastcancer,andmayalterindividualsusceptibilitytocancer.
WeinvestigatedassociationsbetweenmiRNArelatedSNPsandbreastcancerrisk.
Firstweevaluated2,196SNPsinacase-controlstudycombiningninegenomewideassociationstudies(GWAS).
Second,wefurtherinvestigated42SNPswithsuggestiveevidenceforassociationusing41,785casesand41,880controlsfrom41studiesincludedintheBreastCancerAssociationConsortium(BCAC).
CombiningtheGWASandBCACdatawithinameta-analysis,weestimatedmaineffectsonbreastcancerriskaswellasrisksforestrogenreceptor(ER)andagedefinedsubgroups.
FivemiRNAbindingsiteSNPsassociatedsignificantlywithbreastcancerrisk:rs1045494(oddsratio(OR)0.
92;95%confidenceinterval(CI):0.
88–0.
96),rs1052532(OR0.
97;95%CI:0.
95–0.
99),rs10719(OR0.
97;95%CI:0.
94–0.
99),rs4687554(OR0.
97;95%CI:0.
95–0.
99,andrs3134615(OR1.
03;95%CI:1.
01–1.
05)locatedinthe39UTRofCASP8,HDDC3,DROSHA,MUSTN1,andMYCL1,respectively.
DROSHAbelongstomiRNAmachinerygenesandhasacentralroleininitialmiRNAprocessing.
Theremaininggenesareinvolvedindifferentmolecularfunctions,includingapoptosisandgeneexpressionregulation.
FurtherstudiesarewarrantedtoelucidatewhetherthemiRNAbindingsiteSNPsarethecausativevariantsfortheobservedriskeffects.
Citation:KhanS,GrecoD,MichailidouK,MilneRL,MuranenTA,etal.
(2014)MicroRNARelatedPolymorphismsandBreastCancerRisk.
PLoSONE9(11):e109973.
doi:10.
1371/journal.
pone.
0109973Editor:ZhongmingZhao,VanderbiltUniversityMedicalCenter,UnitedStatesofAmericaReceivedJune6,2014;AcceptedSeptember8,2014;PublishedNovember12,2014Thisisanopen-accessarticle,freeofallcopyright,andmaybefreelyreproduced,distributed,transmitted,modified,builtupon,orotherwiseusedbyanyoneforanylawfulpurpose.
TheworkismadeavailableundertheCreativeCommonsCC0publicdomaindedication.
DataAvailability:Theauthorsconfirmthat,forapprovedreasons,someaccessrestrictionsapplytothedataunderlyingthefindings.
DataareavailableviatheBreastCancerAssociationConsortium(BCAC)DataAccessCoordinationCommittee(DACC)(http://ccge.
medschl.
cam.
ac.
uk/consortia/bcac/).
Torequestthedata,readersmaycontactManjeetHumphreys(mkh39@medschl.
cam.
ac.
uk)orDouglasEaston(dfe20@medschl.
cam.
ac.
uk).
Funding:FundingfortheiCOGSinfrastructurecamefromtheEuropeanCommunity'sSeventhFrameworkProgrammeundergrantagreementnumber223175(HEALTH-F2-2009-223175)(COGS).
iCOGSwasalsopartlysupportedbytheCanadianInstitutesofHealthResearchforthe''CIHRTeaminFamilialRisksofBreastCancer''program(JS&DFE),andtheMinistryofEconomicDevelopment,InnovationandExportTradeofQuebec–grant#PSR-SIIRI-701(JS&DFE,P.
Hall).
HEBCSwasfinanciallysupportedbytheHelsinkiUniversityCentralHospitalResearchFund,AcademyofFinland(266528),theFinnishCancerSociety,TheNordicCancerUnionandtheSigridJuseliusFoundation.
ThepopulationalleleandgenotypefrequencieswereobtainedfromthedatasourcefundedbytheNordicCenterofExcellenceinDiseaseGeneticsbasedonsamplesregionallyselectedfromFinland,SwedenandDenmark.
TheUK2GWASwasfundedbyWellcomeTrustandCancerResearchUK.
ItincludedsamplescollectedthroughtheFBCSstudywhichisfundedbyCancerResearchUK[C8620/A8372].
TheWTCCCwasfundedbytheWellcomeTrust.
TheABCFSandOFBCRstudiesweresupportedbytheUnitedStatesNationalCancerInstitute,NationalInstitutesofHealth(NIH)underRFA-CA-06-503andthroughcooperativeagreementswithmembersoftheBreastCancerFamilyRegistry(BCFR)andPrincipalInvestigators,includingCancerCareOntario(U01CA69467),NorthernCaliforniaCancerCenter(U01CA69417),UniversityofMelbourne(U01CA69638).
SamplesfromtheNC-BCFRwereprocessedanddistributedbytheCoriellInstituteforMedicalResearch.
OFBCRwassupportedbytheCanadianInstitutesofHealthResearchforthe''CIHRTeaminFamilialRisksofBreastCancer''programandgrantUM1CA164920fromtheNationalCancerInstitute.
ThecontentofthismanuscriptdoesnotnecessarilyreflecttheviewsorpoliciesoftheNationalCancerInstituteoranyofthecollaboratingcentersintheBreastCancerFamilyRegistry(BCFR),nordoesmentionoftradenames,commercialproducts,ororganizationsimplyendorsementbytheUSGovernmentortheBCFR.
TheABCFSwasalsosupportedbytheNationalHealthandMedicalResearchCouncilofAustralia,theNewSouthWalesCancerCouncil,theVictorianHealthPromotionFoundation(Australia)andtheVictorianBreastCancerResearchConsortium.
JLHisaNationalHealthandMedicalResearchCouncil(NHMRC)AustraliaFellowandaVictorianBreastCancerResearchConsortiumGroupLeader.
MCSisaNHMRCSeniorResearchFellowandaVictorianBreastCancerResearchConsortiumGroupLeader.
JLHandMCSarebothgroupleadersoftheVictoriaBreastCancerResearchConsortium.
TheABCSstudywassupportedbytheDutchCancerSociety[grantsNKI2007-3839;20094363];BBMRI-NL,whichisaResearchInfrastructurefinancedbytheDutchgovernment(NWO184.
021.
007);andtheDutchNationalGenomicsInitiative.
TheBBCSisfundedbyCancerResearchUKandBreakthroughBreastCancerandacknowledgesNHSfundingtotheNIHRBiomedicalResearchCentre,andtheNationalCancerResearchNetwork(NCRN).
TheBBCSGWASreceivedfundingfromTheInstitutNationaldeCancer.
TheworkoftheBBCCwaspartlyfundedbyELAN-FondoftheUniversityHospitalofErlangen.
ES(BIGGS)issupportedbyNIHRComprehensiveBiomedicalResearchCentre,Guy's&St.
Thomas'NHSFoundationTrustinpartnershipwithKing'sCollegeLondon,UnitedKingdom.
ITissupportedbytheOxfordBiomedicalResearchCentre.
TheBSUCHstudywassupportedbytheDietmar-HoppFoundation,theHelmholtzSocietyandtheGermanCancerResearchCenter(DKFZ).
TheCECILEstudywasfundedbyFondationdeFrance[contractgrantnumber2004012618and2007005156],InstitutNationalduCancer(INCa)[2007-1/SPC2,2008-1-CP-4and2009-1-SHS/SP-04],LigueNationalecontreleCancer,AssociationpourlaRecherchecontreleCancer(ARC)[2008-1-CP-4];AgenceFrancaisedeSecuriteSanitairedel'EnvironnementetduTravail(AFSSET-ANSES)[ST-2005-003,EST2008/1/26,andVS-2009-21],LiguecontreleCancerGrandOuest.
TheCGPSwassupportedbytheChiefPhysicianJohanBoserupandLiseBoserupFund,theDanishMedicalResearchCouncilandHerlevHospital.
TheCNIO-BCSwassupportedbytheGenomeSpainFoundation,theRedTematicadeInvestigacionCooperativaenCancerandgrantsfromtheAsociacionEspanolaContraelCancerandtheFondodeInvestigacionSanitario(PI11/00923andPI081120).
WeacknowledgethesupportofAlvarezlvarez,DanielHerrero,PrimitivaMenendezandtheHumanGenotyping-CEGENUnit(CNIO).
TheHumanGenotyping-CEGENUnitissupportedbytheInstitutodeSaludCarlosIII.
TheCTSwassupportedbytheCaliforniaBreastCancerActof1993;NationalInstitutesofHealth(grantsR01CA77398andtheLonVSmithFoundation[LVS39420].
);andtheCaliforniaBreastCancerResearchFund(contract97-10500).
CollectionofcancerincidencedatausedinthisstudywassupportedbytheCaliforniaDepartmentofPublicHealthaspartofthestatewidecancerreportingprogrammandatedbyCaliforniaHealthandSafetyCodeSection103885.
DEMOKRITOSissupportedbyaHellenicCooperativeOncologyGroupresearchgrant(HRR_BG/04)andtheGreekGeneralSecretaryforResearchandTechnology(GSRT)Program,ResearchExcellenceII,fundedat75%bytheEuropeanUnion.
TheDFBBCSGWASwasfundedbyTheNetherlandsOrganisationforScientificResearch(NWO)aspartofaZonMw/VIDIgrantnumber91756341.
ThegenerationandmanagementofGWASgenotypedatafortheRotterdamStudyissupportedbytheNetherlandsOrganisationofScientificResearchNWOInvestments(nr.
175.
010.
2005.
011,911-03-012).
ThisstudyisfundedbytheResearchInstituteforDiseasesintheElderly(014-93-015;RIDE2),theNetherlandsGenomicsInitiative(NGI)/NetherlandsOrganisationforScientificResearch(NWO)projectnr.
050-060-810.
TheRotterdamStudyisfundedbyErasmusMedicalCenterandErasmusUniversity,Rotterdam,NetherlandsOrganizationfortheHealthResearchandDevelopment(ZonMw),theResearchInstituteforDiseasesintheElderly(RIDE),theMinistryofEducation,CultureandScience,theMinistryforHealth,WelfareandSports,theEuropeanCommission(DGXII),andtheMunicipalityofRotterdam.
TheESTHERstudywassupportdbyagrantfromtheBadenWu¨rttembergMinistryofScience,ResearchandArts.
AdditionalcaseswererecruitedinthecontextoftheVERDIstudy,whichwassupportedbyagrantfromtheGermanCancerAid(DeutscheKrebshilfe).
TheHMBCSwassupportedbyagrantfromtheFriendsofHannoverMedicalSchoolandbytheRudolfBartlingFoundation.
TheFinancialsupportforKARBACwasprovidedthroughtheregionalagreementonmedicaltrainingandclinicalresearch(ALF)betweenStockholmCountyCouncilandKarolinskaInstitutet,TheSwedishCancerSocietyandBertvonKantzowfoundation.
TheGC-HBOCwassupportedbyDeutscheKrebshilfe[107054],theDietmar-HoppFoundation,theHelmholtzsocietyandtheGermanCancerResearchCentre(DKFZ).
TheGC-HBOCGWASwasPLOSONE|www.
plosone.
org3November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRisksupportedbytheGermanCancerAid(grantno.
107352).
TheGENICAwasfundedbytheFederalMinistryofEducationandResearch(BMBF)Germanygrants01KW9975/5,01KW9976/8,01KW9977/0and01KW0114,theRobertBoschFoundation,Stuttgart,DeutschesKrebsforschungszentrum(DKFZ),Heidelberg,GermanSocialAccidentInsurance,InstituteoftheRuhrUniversityBochum(IPA),Germany,aswellastheDepartmentofInternalMedicine,EvangelischeKlinikenBonngGmbH,JohanniterKrankenhaus,Bonn,Germany.
TheKBCPwasfinanciallysupportedbythespecialGovernmentFunding(EVO)ofKuopioUniversityHospitalgrants,CancerFundofNorthSavo,theFinnishCancerOrganizations,andbythestrategicfundingoftheUniversityofEasternFinland.
kConFabissupportedbyagrantfromtheNationalBreastCancerFoundation,andpreviouslybytheNationalHealthandMedicalResearchCouncil(NHMRC),theQueenslandCancerFund,theCancerCouncilsofNewSouthWales,Victoria,TasmaniaandSouthAustralia,andtheCancerFoundationofWesternAustralia.
ThekConFabClinicalFollowUpStudywasfundedbytheNHMRC[145684,288704,454508].
FinancialsupportfortheAOCSwasprovidedbytheUnitedStatesArmyMedicalResearchandMaterielCommand[DAMD17-01-1-0729],theCancerCouncilofTasmaniaandCancerFoundationofWesternAustraliaandtheNHMRC[199600].
GCTissupportedbytheNHMRC.
LMBCissupportedbythe'StichtingtegenKanker'(232-2008and196-2010).
DietherLambrechtsissupportedbytheFWOandtheKULPFV/10/016-SymBioSysII.
TheMARIEstudywassupportedbytheDeutscheKrebshilfee.
V.
[70-2892-BRI],theHamburgCancerSociety,theGermanCancerResearchCenterandthegenotypeworkinpartbytheFederalMinistryofEducationandResearch(BMBF)Germany[01KH0402].
MBCSGissupportedbygrantsfromtheItalianAssociationforCancerResearch(AIRC)andbyfundsfromtheItaliancitizenswhoallocatedthe5/1000shareoftheirtaxpaymentinsupportoftheFondazioneIRCCSIstitutoNazionaleTumori,accordingtoItalianlaws(INT-Institutionalstrategicprojects''561000'').
TheMCBCSwassupportedbytheNIHgrants[CA122340,CA128978]andaSpecializedProgramofResearchExcellence(SPORE)inBreastCancer[CA116201],theBreastCancerResearchFoundationandagenerousgiftfromtheDavidF.
andMargaretT.
GrohneFamilyFoundationandtheTingTsungandWeiFongChaoFoundation.
MCCScohortrecruitmentwasfundedbyVicHealthandCancerCouncilVictoria.
TheMCCSwasfurthersupportedbyAustralianNHMRCgrants209057,251553and504711andbyinfrastructureprovidedbyCancerCouncilVictoria.
TheMECwassupportedbyNIHgrantsCA63464,CA54281,CA098758andCA132839.
FortheMTLGEBCSstudy,theinitialcase–controlstudywassupportedbytheCanadianBreastCancerResearchInitiative.
WorkwasalsosupportedbytheQuebecBreastCancerFoundation,theCanadianInstitutesofHealthResearchforthe''CIHRTeaminFamilialRisksofBreastCancer''program–grant#CRN-87521andtheMinistryofEconomicDevelopment,InnovationandExportTrade–grant#PSR-SIIRI-701.
TheNBCSwassupportedbygrantsfromtheNorwegianResearchcouncil,155218/V40,175240/S10toALBD,FUGE-NFR181600/V11toVNKandaSwizzBridgeAwardtoALBD.
TheOBCSwassupportedbyresearchgrantsfromtheFinnishCancerFoundation,theAcademyofFinland,theUniversityofOulu,andtheOuluUniversityHospital.
TheORIGOstudywassupportedbytheDutchCancerSociety(RUL1997-1505)andtheBiobankingandBiomolecularResourcesResearchInfrastructure(BBMRI-NLCP16).
TheOSUstudywasfundedbytheStefanieSpielmanfundandtheOSUComprehensiveCancerCenter.
ThePBCSwasfundedbyIntramuralResearchFundsoftheNationalCancerInstitute,DepartmentofHealthandHumanServices,USA.
ThepKARMAstudywassupportedbyMa¨ritandHansRausingsInitiativeAgainstBreastCancerandCancerRiskPredictionCenter,aLinneusCentre(contract70867902)financedbytheSwedishResearchCouncil.
TheRBCSwasfundedbytheDutchCancerSociety(DDHK2004-3124,DDHK2009-4318).
TheRPCIstudywassupportedbyRPCIDataBankandBioRepository(DBBR),aCancerCenterSupportGrantSharedResource(P30CA016056-32).
TheSASBACstudywassupportedbyfundingfromtheAgencyforScience,TechnologyandResearchofSingapore(A*STAR),theUSNationalInstituteofHealth(NIH)andtheSusanG.
KomenBreastCancerFoundation.
TheSBCSwassupportedbyYorkshireCancerResearchS295,S299,S305PA.
SEARCHisfundedbyaprogrammegrantfromCancerResearchUK[C490/A10124]andsupportedbythetheUKNationalInstituteforHealthResearchBiomedicalResearchCentreattheUniversityofCambridge.
AMDhasbeensupportedbyCancerResearchUKgrant[C8197/A10865]andbytheJosephMitchellFund.
SKKDKFZSissupportedbytheDKFZ.
TheSZBCSwassupportedbyGrantPBZ_KBN_122/P05/2004;KatarzynaJaworskaisafellowofInternationalPhDprogram,PostgraduateSchoolofMolecularMedicine,WarsawMedicalUniversity,supportedbythePolishFoundationofScience.
TheTNBCCwassupportedbytheNIHgrant[CA128978],theBreastCancerResearchFoundation,KomenFoundationfortheCure,theOhioStateUniversityComprehensiveCancerCenter,theStefanieSpielmanfundforBreastCancerResearchandagenerousgiftfromtheDavidF.
andMargaretT.
GrohneFamilyFoundationandtheTingTsungandWeiFongChaoFoundation.
PartoftheTNBCC(DEMOKRITOS)hasbeenco-financedbytheEuropeanUnion(EuropeanSocialFund–ESF)andGreeknationalfundsthroughtheOperationalProgram"EducationandLifelongLearning"oftheNationalStrategicReferenceFramework(NSRF)-ResearchFundingProgramoftheGeneralSecretariatforResearch&Technology:ARISTEIA.
TheUKBGSisfundedbyBreakthroughBreastCancerandtheInstituteofCancerResearch(ICR).
ICRacknowledgesNHSfundingtotheNIHRBiomedicalResearchCentre.
CGEMS,TheNurses'HealthStudiesaresupportedbyNIHgrantsCA65725,CA87969,CA49449,CA67262,CA50385and5UO1CA098233.
Thefundershadnoroleinstudydesign,datacollectionandanalysis,decisiontopublish,orpreparationofthemanuscript.
CompetingInterests:Theauthorshavedeclaredthatnocompetinginterestsexist.
*Email:heli.
nevanlinna@hus.
fi"MembershipoftheGENICANetwork,kConFabInvestigators,andAOCSisprovidedintheAcknowledgments.
IntroductionBreastcanceristhemostcommonwomen'scancerandisaleadingcauseofcancermortality[1].
Inheritedgeneticvariationhasbeenassociatedwiththeinitiation,developmentandprogressionofbreastcancer.
Studiesontwinshavesuggestedthathereditarypredisposingfactorsareinvolvedinuptoonethirdofallbreastcancers[2].
Manygeneticlocihavebeenassociatedwithbreastcancerriskandcollectivelyexplainapproximately35%ofthefamilialrisk[3,4].
Thelargestgeneticassociationstudyofbreastcancertodateidentified41novellowpenetrancesusceptibilityloci[4]byselectingnearly30,000SNPsfromameta-analysisofninegenome-wideassociation(GWA)studiesandgenotypingthemusing41,785casesand41,880controlsofEuropeanancestryfromstudiesintheBreastCancerAssociationConsortium(BCAC).
These41susceptibilitylociprobablyrepresentthetipoftheiceberg,andadditionalSNPsfromthecombinedGWASmightexplainasimilarfractionoffamilialrisktothatattributedtothealreadyidentifiedloci[4].
MaturemiRNAsare20–23nucleotide,single-strandedRNAmoleculesthatplayacrucialroleingeneexpressionregulationformanycellularprocessesincludingdifferentiationpotentialanddevelopmentpattern.
MiRNAsundergoastepwisematurationprocessinvolvinganarrayofmiRNAmachinerycomponents.
DroshaandDGCR8mediatethecleavageoflongprimarymiRNAtranscripts(pri-miRNAs)intoshorterpre-miRNAsinthenucleus[5,6].
Thepre-miRNAsarethentransportedtothecytoplasmwheretheyarefurthercleavedbyDicertoproducematuremiRNAs[7].
MiRNAsinteractbypairingwiththe39untranslatedregion(UTR),andalsowithinthecodingregionand59UTRofthecorrespondingmRNAsleadingtomRNAdestabilization,cleavageortranslationrepression.
MoreeffectivemRNAdestabilizationisachievedwhenmiRNAtargetsthe3'UTRratherthanothermRNAregions[8–10].
AnindividualmiRNAmayregulateapproximately100distinctmRNAs,andtogethermorethan1000humanmiRNAsarebelievedtomodulatemorethanhalfofthemRNAspeciesencodedinthegenome[11,12].
Additionally,mostmRNAspossessbindingsitesformiRNAs[13].
MiRNAsareinvolvedintumorigenesisinthattheycanbeeitheroncogenicwhentumorsuppressorgenesaretargeted,orgenomicguardians(tumoursuppressormiRNAs)whenoncogenesaretargeted[14].
Additionallyithasbeensuggestedthattheymaymodulatebothmetastasis[15]andchemotherapyresistance[16].
MiRNAshavealsobeenshowntohavealteredexpressionlevelsintumourscomparedtonormaltissueandbetweentumorsubtypesinbreastcanceramongothercarcinomatypes[17–19].
SNPsmayaffectmiRNAmachinerygenesormiRNAsactivity;howeverSNPscanalsocreate,abolishormodifymiRNAbindingsitesintheirbindingregions.
PolymorphismsinmiRNAbindingsiteshavebeenstudiedinregardtotheriskofseveralcancers[20],includingbreastcancer[21–23].
ThesestudieshavefoundevidenceforassociationofmiRNArelatedSNPsandcancerrisk,butthestudysamplesizeshavebeenrelativelysmall.
Inthisstudy,weinvestigateassociationsbetweenmiRNA-relatedpolymorphismsandbreastcancerriskbyusingameta-PLOSONE|www.
plosone.
org4November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRiskanalysisofnineGWASandsubsequentgenotypingoftophitsusing41,785casesand41,880controlsofEuropeanancestryfromtheBCAC.
Toourknowledge,thisisthusfarthelargestinvestigationofassociationsbetweenmiRNA-relatedpolymor-phismsandbreastcancersusceptibility.
MaterialsandMethodsSNPselectionandgenotypingSNPsinmatureorpre-miRNAs,ingenesofthemiRNAmachineryandin3'UTRregionsofproteincodinggeneswithapotentialeffectonmiRNAbindingweresystematicallysearchedfromEnsembl(hg18/build36)andPatroclesdatabases[24].
Additionally,taggingSNPsforsuchwithr2$0.
8werealsoidentifiedutilizingthepublicHapMapSNPdatabase.
Bythisinsilicoapproachweidentifiedaltogether147,801candidateSNPsand12,550taggingSNPs.
TheseSNPswerethenoverlayedwiththosefromthecombinedGWASfromtheBCAC[4]andaltogether2196SNPswerepresent(eithergenotypedorimputed)inthecombinedGWAS.
TheseSNPsweregenotypedwithIlluminaorAffymetrixarrays,asdescribedpreviously[25–32].
ThecombinedGWASdatawereimputedforallscansusingHapMapversion2CEUasareferenceinsimilarfashiontothatpresentedbyMichailidouandcolleagues[4]withtheexceptionthattheHapMapversion2release21wasusedatthetimetheoverlaywasperformed.
Analysisusinga1-degree-of-freedomtrendtestofthese2196SNPsinthecombinedGWASindicatedsomeevidenceofassociationwithbreastcancerriskfor44SNPs(p,0.
09).
Notably,thecombinedGWASincludedimputeddatageneratedusingHapMapversion2release21(basedonNCBIbuild35(dbSNPb125)),whereastheresultspresentedhereforthecombinedGWASarebasedonimputationusingHapMapversion2release22(basedonNCBIbuild36(dbSNPb126)).
Intherelease22,anumberofSNPswereexcludedduetomappinginconsistenciesinbuild35relativetobuild36.
Hence,theestimatesfromthecombinedGWASmayslightlydifferfromtheinitialassociationanalysis.
The44SNPs(including30candidateand14taggingSNP)weregenotypedonadditionalsamplesintheBCACusingthecustomIlluminaInfiniumarray(iCOGS)whichincludedatotalof211,155SNPsasdescribedpreviously.
ThedetaileddescriptionofqualitycontrolprocessforcombinedGWASandiCOGSgenotypingdatawaspresentedin[4].
Ofthe42SNPsthatpassedqualitycontrol[4],twowerelocatedinmiRNAgenes(onecandidateSNPlocatedinpre-miRNAhsa-miR-2110andonetagSNPtaggingamaturehsa-mir-548lvariant),andfourSNPswerelocatedinmiRNAmachinerygenes(SMAD5,SND1,CNOT4andDROSHA).
ThegenotypedDROSHASNPtagsthe39UTRmiRNAbindingsitevariantintheDROSHAgene.
Theremaining38candidateortagSNPswerelocatedin,ortaggedtoapredictedmiRNAbindingsiteinthe39UTRofproteincodinggenes.
All42SNPsaredescribedinTable1.
TheworkflowoftheSNPselectionindifferentstagesisillustratedinFigure1.
StudysampleThecombinedGWASincludedninebreastcancerstudiestotalling10,052casesand12,575controlsofEuropeanethnicbackground.
Detailsandstudy-specificsubjectnumbersarepresentedinTableS1.
SincetheGWASwerelimitedtopatientsofEuropeanethnicbackgroundwefurtherutilized41,785casesascertainedfortheirfirstprimary,invasivebreastcancerand41,880controlsofEuropeanancestryfrom41BCACstudiesgenotypedusingtheiCOGSarray(TableS2).
ForasubgroupanalysisofERnegativeandERpositivecases,aswellascasesagedlessthan50yearsatdiagnosis,weincludedallthecasesforwhichtherespectivedatawereavailable.
TheERsubgroupanalysiswasbasedon702ERnegativecasesand2,019ERpositivecasesfromfiveGWASstudiesand7,200ERnegativecasesfrom40BCACstudiesand26,302ERpositivecasesfrom34BCACstudies.
Theanalysisofcasesagedlessthan50yearsatdiagnosiswasbasedon3,470casesfromthreeGWASstudiesand9,483casesfrom35BCACstudies.
AllparticipatingstudiesconformtotheDeclarationofHelsinkiandwereapprovedbytherespectiveethicalreviewboardsandethicscommittees(TablesS1andS2),andallparticipantsinthesestudieshadprovidedwrittenconsentfortheresearch.
StatisticalmethodsWeusedlogisticregressiontoestimateper-allelelog-oddsratiosandstandarderrorsincludingthestudyasacovariate.
Wealsoincludedprincipalcomponentsascovariatesinordertocorrectforpotentialhiddenpopulationstructure.
IntheGWAS,fortwostudies(UK2andHEBCS)theestimateswereadjustedforthefirstthreeprincipalcomponentsandintheiCOGSanalysisweusedthefirstsixprincipalcomponentsandanadditionalcomponenttoreduceinflationfortheLMBCstudy,asdescribedpreviously[4].
SubgroupanalyseswerecarriedoutforERnegativeandpositivesubgroupsandforthegroupagedlessthan50yearsatdiagnosis.
Formeta-analysis,wecombinedtheestimatesfromthecombinedGWASandiCOGSwithafixedeffectsmodelusingtheinversevarianceweightedmethod.
Inthemeta-analysis,thesubjectsinvolvedinbothcombinedGWASandiCOGS(1880)wereonlytakenintoaccountonce.
InordertoadustforP-valuesagainstmultipletesting,weusedBenjaminiHochbergcorrection.
TheadjustedP-valuesareshowninTable2alongwiththenominalP-values.
InthetextwereportthenominalP-values.
ThestatisticalanalyseswereconductedusingtheR2.
14.
0statisticalcomputingenvironment(http://www.
r-project.
org/).
ResultsForthe42SNPswesuccessfullygenotyped,estimatesofassociationfromthecombinedGWASandfromiCOGSanalysisareshowninTableS3.
Twenty-oneSNPsshowedconsistentFigure1.
WorkflowofmiRNASNPselection.
doi:10.
1371/journal.
pone.
0109973.
g001PLOSONE|www.
plosone.
org5November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRiskTable1.
The42studiedSNPsinmiRNAs,miRNAmachinerygenesandmiRNAtargetgenes.
FunctionalSNP(TagSNP,R-squared)ChrPositionCodingGenemiRNASNPeffectaLocatedwithinmiRNArs1709140310115923895GAhsa-miR-2110rs13447640(rs1805360,r2=1)1193866677GAhsa-mir-548lLocatedinmiRNAbiogenesismachinerygenesrs37649415135497426ACSMAD5rs171516397127425052AGSND1rs174806167134773600CGCNOT4rs10719531437204GADROSHAhsa-miR-1298ACLocatedinmiRNAtargetgenesrs25503031654953111AGAMFRhsa-miR-577ACrs7513934152590776GACC2D1Bhsa-miR-384/hsa-miR-577CNCrs1128226721908194ACCDCA7Lhsa-miR-548gACrs37961333100000533GADCBLD2hsa-miR-624*ACrs74411290063806GADCNhsa-miR-135b*ACrs18034392137807312AGDYRK1Ahsa-miR-550ACrs37971527199858AGFAM189A1hsa-miR-570ACrs713062211128186721ACFLI1hsa-miR-138-2*ACrs10525321589275240AGHDDC3hsa-miR-1224-3p/hsa-miR-1260/hsa-miR-1280ACrs704012397160742AGKDM4Chsa-miR-154*/hsa-miR-487aACrs10622251049313232AGMAPK8hsa-miR-203ACrs417397116224740AGMEThsa-miR-576-5pACrs702681556253786AGMIER3hsa-miR-196a*ACrs3134615140134653CAMYCL1hsa-miR-1827ANCrs23046692238830402AGPER2hsa-miR-885-3pACrs134221715074900ACPMP22hsa-miR-29b-1*ACrs75623912201444411ACPPIL3hsa-miR-493*/hsa-miR-499-3pACrs7520333140862837AGRIMS3hsa-let-7d/hsa-let-7eCNCrs7396921853178524GAST8SIA3hsa-miR-96/hsa-miR-1271/hsa-miR-182ACrs10584504120200088GASYNPO2hsa-miR-183ACrs4351800117446395CASYT9hsa-miR-544ACrs124383241555366808AGTCF12hsa-miR-591ACrs128698701399415306GAZIC5hsa-miR-34a/hsa-miR-34c-5p/hsa-miR-449a/hsa-miR-449bACrs9990(rs1444418,r2=1)1064230476AGADOhsa-miR-512-5p/hsa-miR-510ACrs757537(rs4705870,r2=1)5132187033GAANKRD43hsa-miR-320a/hsa-miR-320b/hsa-miR-320c/hsa-miR-320dACrs3774729(rs2037119,r2=0.
943)363969919GAATXN7hsa-miR-1206ACPLOSONE|www.
plosone.
org6November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRiskassociationswithbreastcancerriskinthecombinedGWASandiniCOGSanalysis;resultsfromthemeta-analysisareshowninTable2.
ThemostsignificantlyassociatedSNP,rs702681(OR1.
06[95%CI1.
04–1.
08];P3.
9610210),islocatedinthe3'UTRofMIER3,closetotheknownbreastcancersusceptibilitygeneMAP3K1.
TheSNPrs702681islocatedatthesame5q11.
2locusasthepreviouslypublishedriskSNPrs889312[33](correlationr2=0.
3).
WhenthetwoSNPswereanalysedinthesamelogisticregressionmodel,theassociationwithrs889312,butnotthatwithrs702681remainednominallystatisticallysignificant,suggestingthatrs702681isunlikelytobethecausalSNPatthislocus.
ThefiveSNPswiththesignificantnovelassociationsfromthemeta-analysis(P#5.
0761023andadjustedP#3.
5561022aftercorrec-tionformultipletesting)werers1045494,(OR0.
92[95%CI0.
88–0.
96];P=5.
9061025),rs1052532,(OR0.
97[95%CI0.
95–0.
99];P=7.
7861024),rs10719,(OR0.
97[95%CI0.
94–0.
99];P=1.
3561023)rs4687554(OR0.
97[95%CI0.
95–0.
99];P=1.
7161023)andrs3134615(OR1.
03[95%CI1.
01–1.
05];P=5.
0761023)locatedin39UTRofCaspase-8(CASP8),HDDomainContaining3(HDDC3),DROSHA,Musculoskeletal,EmbryonicNuclearProtein1(MUSTN1)andV-MycMyelocy-tomatosisViralOncogeneHomolog1(MYCL1),respectively(Table2).
SNPrs1045494istaggingthehsa-miR-938bindingsiteSNPrs1045487(r2=1.
0)ofCASP8andtheSNPrs1052532inHDDC3ispredictedtoabolishthebindingsiteforhsa-miR-1224-3p.
TheSNPrs10719ispredictedtoabolishthehsa-miR-1298bindingsiteinthe39UTRofDROSHA.
SNPrs4687554tagsthehsa-miR-891bbindingsiteSNPrs6445538(r2=1.
0)ofMUSTN1andrs3134615islocatedatthebindingsiteofhsa-miR-1827ofMYCL1.
Therewasnoevidenceforheterogeneityintheper-alleleORforanySNP.
TheperstudyperalleleORsforthesefivemiRNAbindingsiteSNPsfromthecombinedGWASalongwithper-SNPheterogeneityvarianceP-valuesareshowninFigureS1andfromtheiCOGSinFigureS2.
NextweanalysedtheSNPsbyERstatus-definedsubtype,andforcasesagedlessthan50yearsatdiagnosis,forriskassociationsinthemeta-analysisofcombinedGWASandiCOGS(TablesS4,S5andS6).
Theseanalysesdidnotrevealanyadditionalsignificantresults.
Forrs1045494inCASP8,rs4687554inMUSTN1andrs3134615inMYCL1(OR1.
03[95%CI1.
01–1.
05];P=7.
7561024)amoresignificantassociationwithbreastcancerriskwasfoundfortheERpositivesubgroupthaninthemainanalysis,buttheresultfromthetestforheterogeneitybyERstatuswasnotsignificant(datanotshown).
Allassociationswereestimatedusinganadditiveinheritancemodel.
Dominantandrecessivemodelsdidnotimprovetheestimates(datanotshown).
DiscussionWeinvestigatedassociationsbetweengeneticvariationinmiRNAs,inthegenesofthemiRNAmachineryandinthemiRNAbindingsitesandtheriskofbreastcancer.
WeidentifiedseveralSNPsthatarepredictedtoabolishanmiRNAbindingsiteandthataresignificantlyassociatedwithbreastcancerrisk.
PreviousstudiesinvestigatingmiRNArelatedSNPs,especiallyinmiRNAbindingsiteshaveincludedpredefinedsetsofgenes.
Nicolosoandcolleaguesinvestigated38previouslyidentifiedbreastcancerriskSNPsandfoundtwotomodifymiRNAbindingsitesinTGFB1andXRCC1invitro[23].
Neitherofthesewereincludedinourdataset.
Liangandcolleaguesinvestigated134potentialmiRNAbindingsitesincancer-relatedgenesandfoundsixmiRNAbindingsiteSNPsthatwereassociatedwithovariancancerrisk[34].
Table1.
Cont.
FunctionalSNP(TagSNP,R-squared)ChrPositionCodingGenemiRNASNPeffectars1045487(rs1045494,r2=1)2201860026AGCASP8hsa-miR-938ACrs7288826(rs8140217,r21)2237547947GACBX6hsa-miR-1207-5pACrs17569034(rs17512204,r2=0.
835)2118449301GACCDC93hsa-miR-1178ACrs3205281(rs7674744,r2=1)478874296GACNOT6Lhsa-miR-643/hsa-miR-297ACrs13005(rs9473,r2=0.
964)1013727177GAFRMD4Ahsa-miR-548mACrs3809831(rs3809828,r2=1)177187575GAKCTD11hsa-miR-892bACrs6445538(rs4687554,r2=1)352839175AGMUSTN1hsa-miR-891bACrs7818(rs9371201,r2=0.
875)6150186694GAPCMT1hsa-miR-595ACrs9844202(rs7635553,r2=1)3168646064GASERPINI2hsa-miR-1272ACrs2271565(rs7086917,r2=1)1049867441ACWDFY4hsa-miR-657/hsa-miR-214/hsa-miR-15a/hsa-miR-16/hsa-miR-15b/hsa-miR-195/hsa-miR-424/hsa-miR-497ACTagSNPsusedintheanalysisarepresentedintheparenthesisalongwiththeRsquredvaluerelativetothefunctionalSNP.
aAccordingtoPatroclesprediction;AC=abolishesconservedbindingsite,ANC=abolishesnon-conservedbindingsite,CNC=createsnon-conservedbindingsite(Targetsitesareconsideredconservediftheyaresharedbyatleastoneprimate,onerodentandonenonprimate/nonrodentmammal[24]).
doi:10.
1371/journal.
pone.
0109973.
t001PLOSONE|www.
plosone.
org7November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRiskTable2.
AssociationsofSNPsintheGWASandiCOGSseparatelyandcombinedGWAS+iCOGSandbreastcancerrisk.
SNPChrPositioncoding1GWASOR(95%CI)2GWASP3iCOGSOR(95%CI)2iCOGSP3CombinedGWAS+iCOGSOR(95%CI)2CombinedGWAS+iCOGSP3(BHcorrectedP)4Geners702681556253786AG1.
07(1.
02–1.
11)3.
92610231.
06(1.
04–1.
09)2.
76610281.
06(1.
04–1.
08)3.
88610210(1.
6361028)MIER3rs10454942201860026AG0.
90(0.
81–1.
00)4.
74610220.
92(0.
88–0.
96)4.
47610240.
92(0.
88–0.
96)5.
9461025(1.
2561023)CASP8rs10525321589275240AG0.
94(0.
90–0.
98)7.
94610230.
97(0.
95–0.
99)1.
47610220.
97(0.
95–0.
99)7.
7861024(1.
0961022)HDDC3rs10719531437204GA0.
92(0.
88–0.
97)8.
79610240.
98(0.
95–1.
00)5.
32610220.
97(0.
94–0.
99)1.
3561023(1.
4261022)DROSHArs4687554352839175AG0.
94(0.
90–0.
99)1.
23610220.
97(0.
95–1.
00)2.
39610220.
97(0.
95–0.
99)1.
7161023(1.
4461022)MUSTN1rs3134615140134653CA1.
04(0.
99–1.
09)9.
97610221.
03(1.
00–1.
05)2.
09610221.
03(1.
01–1.
05)5.
0761023(3.
5561022)MYCL1rs76355533168646064GA0.
89(0.
83–0.
95)9.
73610240.
98(0.
95–1.
01)1.
98610211.
00(0.
97–1.
04)9.
2461023(5.
5461022)SERPINI2rs37961333100000533GA1.
18(1.
08–1.
29)4.
18610241.
01(0.
97–1.
06)5.
74610211.
04(1.
00–1.
09)3.
9361022(1.
4561021)DCBLD2rs4351800117446395CA1.
04(1.
00–1.
08)4.
48610221.
01(0.
99–1.
03)1.
98610211.
02(1.
00–1.
04)4.
1561022(1.
4561021)SYT9rs175122042118449301GA1.
06(0.
98–1.
14)1.
20610211.
03(0.
99–1.
06)1.
63610211.
03(1.
00–1.
07)5.
2261022(1.
5761021)CCDC93rs3809828177187575GA1.
17(1.
06–1.
28)1.
97610231.
01(0.
97–1.
05)5.
22610210.
99(0.
95–1.
03)7.
9361022(2.
2261021)KCTD11rs74411290063806GA1.
11(1.
03–1.
20)8.
70610231.
01(0.
97–1.
05)5.
98610211.
03(0.
99–1.
06)1.
0461021(2.
5761021)DCNrs70869171049867441AC0.
96(0.
93–1.
00)6.
35610220.
99(0.
97–1.
01)4.
38610210.
99(0.
97–1.
00)1.
2961021(3.
0161021)WDFY4rs704012397160742AG1.
11(0.
99–1.
23)7.
59610221.
02(0.
97–1.
07)5.
14610211.
00(0.
95–1.
04)1.
7961021(3.
7461021)KDM4Crs7674744478874296GA0.
94(0.
89–0.
99)2.
83610220.
99(0.
97–1.
02)6.
91610211.
01(0.
98–1.
03)1.
8161021(3.
7461021)CNOT6Lrs124383241555366808AG0.
87(0.
79–0.
97)1.
01610221.
00(0.
94–1.
05)8.
69610211.
02(0.
98–1.
07)1.
8761021(3.
7461021)TCF12rs171516397127425052AG0.
96(0.
92–1.
01)1.
09610210.
99(0.
97–1.
02)5.
66610211.
00(0.
98–1.
02)2.
1961021(4.
1861021)SND1rs174806167134773600CG0.
87(0.
72–1.
04)1.
27610210.
99(0.
93–1.
04)6.
39610210.
98(0.
92–1.
03)3.
7061021(5.
9861021)CNOT4rs7513934152590776GA1.
04(1.
00–1.
08)7.
98610221.
00(0.
98–1.
02)9.
99610211.
01(0.
99–1.
02)4.
3761021(6.
3461021)CC2D1Brs23046692238830402AG0.
96(0.
91–1.
02)1.
86610211.
00(0.
97–1.
02)8.
17610210.
99(0.
97–1.
02)4.
3861021(6.
3461021)PER2rs10584504120200088GA0.
96(0.
91–1.
01)1.
33610211.
00(0.
97–1.
02)9.
28610211.
01(0.
98–1.
03)4.
5961021(6.
4361021)SYNPO2TheSNPswithconsistentoddsratiosincombinedGWASandiCOGSanalysisareshown.
(Resultsforall42SNPsarepresentedinTableS3.
)1Build36position.
2Peralleleoddsratiofortheminorallelerelativetothemajorallele.
31dfp-trend.
41dfp-trendadjustedagainstmultipletestingbyBenjamini–Hochbergcorrectionmethod.
doi:10.
1371/journal.
pone.
0109973.
t002PLOSONE|www.
plosone.
org8November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRiskInthemeta-analysisofcombinedGWASandiCOGSformaineffects,forfourofthefivemostsignificantmiRNAbindingsiteSNPs,theminorallelewasassociatedwithadecreasedbreastcancerrisk.
TheminoralleleofSNPrs3134615in39UTRofMYCL1wasassociatedwithanincreasedbreastcancerrisk.
AllthefivemostsignificantmiRNAbindingsiteSNPslocatein39UTRandhavebeenpredictedtoabolishthemiRNAbindingsite.
ThedefectinmiRNA-mediatedregulationwouldbeexpectedtoleadtoanincreaseinthetranslationofthecorrespondingencodedprotein.
Thefivegenes,whoseregulationmaybeaffectedbythemiRNA-associatedSNPs,includethepre-apoptoticgeneCASP8,HDDC3,miRNAbiogenesismasterregulatorDROSHA,MYC-familymemberMYCL1andMUSTN1.
CASP8isinvolvedinapoptosisinbreastcancercells[35],andmanystudieshavereportedpolymorphismsinthisgenetobeassociatedwithrisksforseveralcancers[36,37]includingbreastcancer[38,39],indicatingtheimportanceofCASP8intumordevelopment.
SNPrs1045494studiedhereislocatedclosetothecodingregionSNPrs1045485thathasbeenpreviouslyshowntohaveastrongerprotectiveeffect[38,40,41].
Interestingly,MichalidouandcolleaguesreportedthisSNPashavingonlyweakevidenceforanassociation(P0.
0013incombinedGWASandiCOGS)[4],butthesetwoSNPs(rs1045485andrs1045494)arenotcorrelated(r2=0.
001inCaucasianpopulation).
Neitherisrs1045494correlatedwiththemorestronglyassociatedrs1830298SNP,identifiedthroughfine-mappingoftheregion(r2=0.
02)[42].
Rs1045494tagsSNPrs1045487(r2=1.
0)whichispredictedtoabolishthehsa-miR-938bindingsiteandthusmayaffectCASP8expression.
ThereisverylittlereportedevidenceontheinvolvementofHDDC3orthehsa-miR-1224-3pincancer,indicatinganovelassociationwithrisk.
HDDC3hasbeensuggestedtobeinvolvedinthestarvationresponse[43].
TheHDDC3geneisexpressedathigherlevelsbyseveraldifferenttumortypes,includingbreasttumors,thanbynormaltissue[44].
DROSHAisamiRNAmasterregulator.
ItisamemberoftheRNaseIIIenzymefamily,belongstothemiRNAbiogenesispathwayandisthecorenucleasethatprocessespri-miRNAsintopre-miRNAsinthenucleus[5,6].
TheSNPrs10719inthe39UTRofDROSHAispredictedtoabolishthehsa-miR-1298bindingsite.
Hsa-miR-1298ispredictedtotargetDROSHAbythePatroclespredictionaswellasbyTargetScan[45]andPITA[46]predictionalgorithms.
RecentlyasmallKoreanstudyreportedanotherSNPrs644236,taggingtheSNPrs10719(r2=0.
955inCEUpopulationandr2=0.
876inAsianpopulation(combinedCHBandJPT))tobeassociatedwithelevatedbreastcancerrisk[47].
WhentakingintoaccounttheoppositemajorandminorsallelesintheAsianandEuropeanpopulationsforSNPsrs644236andrs10719,thisresultisinconcordancewithourresultswhereboththecombinedGWASaswellastheiCOGSanalysisconsistentlyindicatedanassociationoftheminoralleleofSNPrs10719withreducedbreastcancerrisk.
WealsofoundtheminoralleleofSNPrs3134615inthe39UTRofMYCL1tobeassociatedwithanincreasedrisk.
MYCL1(L-MYC)belongstothesamefamilyoftranscriptionfactorsastheknownproto-oncogeneMYC(C-MYC)andtheyshareahighdegreeofstructuralsimilarity[48].
TheMYCL1genehaspreviouslybeenreportedtobeamplifiedandoverexpressedinovariancancer[49].
Acase-controlstudybyXiongandcolleaguesreportedSNPrs3134615tobesignificantlyassociatedwithincreasedriskofsmallcelllungcancer[50].
SNPrs3134615waspredictedbyPatroclestoabolishthehsa-miR-1827bindingsite.
ThishasalsobeensuggestedbyfunctionalstudieswhereMYCL1wasfoundasthetargetofhsa-miR-1827andtheSNPrs3134615wasalsofoundtoincreaseMYCL1expression[50].
TheevidencefromfunctionalstudiesisconsistentwithourfindingthatSNPrs3134615mightincreasebreastcancerrisk.
MUSTN1hasbeenshowntobeinvolvedinthedevelopmentandregenerationofthemusculoskeletalsystem[51].
ThusfarnoevidenceofassociationbetweenMUSTN1andbreastcancerhasbeenreported,buttheMUSTN1geneisexpressedinthemammaryglands[52].
SinceonlyasmallfractionofmiRNAbindingsiteshasbeenexperimentallyvalidated,weselectedSNPsthathadbeencomputationallypredictedtoaffectmiRNAbindingsites.
ForouroriginalSNPselectionweusedthePatroclesdatabasethatcontainspredictedmiRNAbindingsitesandalsocompilesperturbationpredictionofSNPeffects.
Thereareamultitudeofpredictionprogramsandtheirperformancehasbeenevaluated[53].
Witkosandcolleaguesfindtargetpredictionalgorithmsthatutilizeorthologoussequencealignment,likePatrocles,tobethemostreliable.
Thefollowupofthe42miRNArelatedSNPsidentifiedfivesignificantassociationswithbreastcancerrisk.
Althoughtheindividualriskeffectsweresubtle,consideringthatwecouldonlyinvestigateasmallproportionofourinitialinsilicodatasetofmiRNArelatedSNPs(over140,000SNPs)thismaysuggestthatgeneticpolymorphismsaffectingthemiRNAregulationcouldhaveaconsiderablecombinedeffectonbreastcancerrisk.
Itshouldbenotedthat,untilfinemappingstudiesarecarriedoutfortheseloci,itisnotclearwhetherthesemiRNA-relatedSNPsarethevariantsresponsiblefortheobservedassociations.
ThiscomprehensiveanalysisofmiRNArelatedpolymorphismsusingalargetwostagestudyofwomenwithEuropeanancestryprovidesevidenceformiRNArelatedSNPsbeingpotentialmodulatorsofbreastcancerrisk.
SupportingInformationFigureS1ForestplotsforthefivemostsignificantmiRNAbindingsiteSNPsfromthecombinedGWAS.
Squaresindicatetheestimatedper-alleleORfortheminoralleleinEuropeans.
Thehorizontallinesindicate95%confidencelimits.
Theverticalbluedashedlinesindicateclippingoftheconfidenceintervalsforpresentationpurpose.
Theareaofthesquareisinverselyproportionaltothevarianceoftheestimate.
Thediamondindicatestheestimatedper-alleleORfromthecombinedanalysis.
(PDF)FigureS2ForestplotsforthefivemostsignificantmiRNAbindingsiteSNPsfromtheiCOGS.
Squaresindicatetheestimatedper-alleleORfortheminoralleleinEuropeans.
Thehorizontallinesindicate95%confidencelimits.
Theverticalbluedashedlinesindicateclippingoftheconfidenceintervalsforpresentationpurpose.
Theareaofthesquareisinverselyproportionaltothevarianceoftheestimate.
Thediamondindicatestheestimatedper-alleleORfromthecombinedanalysis.
(PDF)TableS1AdescriptionofeachGWASstudy,numberofsubjectsandgenotypingplatformusedincombinedGWAS.
(DOC)TableS2AdescriptionofeachBCACstudywithsubjectsofEuropeanorigininiCOGS.
(DOC)TableS3Frequenciesandeffectsizesofthe42SNPsinthemainanalysis;combinedGWASandiCOGS.
(DOC)PLOSONE|www.
plosone.
org9November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRiskTableS4ResultsforSNPsintheGWASandiCOGSseparatelyandcombinedGWAS+iCOGSanalysisforERnegativesubgroup.
(DOC)TableS5ResultsforSNPsintheGWASandiCOGSseparatelyandcombinedGWAS+iCOGSanalysisforERpositivesubgroup.
(DOC)TableS6ResultsforSNPsintheGWASandiCOGSseparatelyandcombinedGWAS+iCOGSanalysisforcaseslessthan50yearsatdiagnosis.
(DOC)AcknowledgmentsWethankalltheindividualswhotookpartinthesestudiesandalltheresearchers,studystaff,cliniciansandotherhealthcareproviders,techniciansandadministrativestaffwhohaveenabledthisworktobecarriedout.
TheHEBCSthanksDr.
KarlvonSmittenandRNIrjaErkkila¨fortheirhelpwiththeHEBCSdataandsamples.
TheABCFSthanksMaggieAngelakos,JudiMaskiellandGillianDite.
TheOFBCRthanksTeresaSelander,NayanaWeerasooriyaandGordGlendon.
TheABCSwouldliketoacknowledgeEllenvanderSchootforDNAofcontrols.
TheBBCCthanksSilkeLandrith,SonjaOeser,MatthiasRu¨bner.
TheBBCSthanksEileenWilliams,ElaineRyder-MillsandKaraSargus.
TheBIGGSthanksNiallMcInerney,GabrielleColleran,AndrewRowanandAngelaJones.
TheBSUCHthanksPeterBugertandtheMedicalFaculty,Mannheim.
TheCGPSthanksthestaffandparticipantsoftheCopenhagenGeneralPopulationStudy,andDortheUldallAndersen,MariaBirnaArnadottir,AnneBank,DortheKjeldgardHansenforexcellenttechnicalassistance.
TheCNIO-BCSacknowledgethesupportofNuriaAlvarez,DanielHerrero,PrimitivaMenendezandtheHumanGenotyping-CEGENUnit(CNIO).
TheDFBBCSthanksMargreetAusems,ChristivanAsperen,SennoVerhoef,andRogiervanOldenburgforprovidingsamplesfromtheirClinicalGeneticcenters.
WealsothankPascalArp,MilaJhamai,MarijnVerkerk,LizbethHerreraandMarjoleinPetersfortheirhelpincreatingtheGWASdatabase,andKarolEstradaandMaksimV.
Struchalinfortheirsupportincreationandanalysisofimputeddata.
Theauthorsaregratefultothestudyparticipants,thestafffromtheRotterdamStudyandtheparticipatinggeneralpractitionersandpharmacists.
TheESTHERthanksHartwigZiegler,SonjaWolfandVolkerHermann,KatjaButterbach.
TheGC-HBOCwouldliketothankthefollowingpersonsforprovidingadditionalinformationandsamples:Prof.
Dr.
NorbertArnold,Dr.
SabinePreissler-Adams,Dr.
MonikaMareeva-Varon,Dr.
DieterNiederacher,Prof.
Dr.
BrigitteSchlegelberger,Dr.
ClemensMu¨l,HeideHellebrand,andStefanieEngert.
TheHMBCSthanksPeterHillemanns,HansChristiansenandJohannH.
Karstens.
TheKBCPthanksEijaMyo¨ha¨nenandHelenaKemila¨inen.
kConFab/AOCSwishtothankHeatherThorne,EvelineNiedermayr,allthekConFabresearchnursesandstaff,theheadsandstaffoftheFamilyCancerClinics,andtheClinicalFollowUpStudyfortheircontributionstothisresource,andthemanyfamilieswhocontributetokConFab.
TheLMBCthanksGilianPeuteman,DominiekSmeets,ThomasVanBrusselandKathleenCorthouts.
TheMARIEwouldliketothankAlinaVrieling,KatharinaBuck,UrsulaEilber,MuhabbetCelik,andSabineBehrens.
TheMBCSGthanksSiranoushManoukian,BernardPeisselandDanielaZaffaronioftheFondazioneIRCCSIstitutoNazionaledeiTumori(INT);BernardoBonanni,IreneFeroceandAngelaManiscalcooftheIstitutoEuropeodiOncologia(IEO)andthepersonneloftheCogentechCancerGeneticTestLaboratory.
TheMTLGEBCSgratefullyacknowledgetheassistanceofLesleyRichardsonandMarie-ClaireGouletinconductingthestudy.
WewouldliketothankMartineTranchant(CancerGenomicsLaboratory,CHUdeQuebecResearchCenter),Marie-FranceValois,AnnieTurgeonandLeaHeguy(McGillUniversityHealthCenter,RoyalVictoriaHospital;McGillUniversity)forDNAextraction,samplemanagementandskillfultechnicalassistance.
J.
S.
isChairholderoftheCanadaResearchChairinOncogenetics.
TheOBCSthanksMeeriOtsukkaandKariMononen.
TheORIGOthanksE.
Krol-Warmerdam,andJ.
Blomforpatientaccrual,administeringquestionnaires,andmanagingclinicalinformation.
TheLUMCsurvivaldatawereretrievedfromtheLeidenhospital-basedcancerregistrysystem(ONCDOC)withthehelpofDr.
J.
Molenaar.
TheOSUthanksRobertPilarksiandCharlesShapiro,whowereinstrumentalintheformationoftheOSUBreastCancerTissueBank.
WethanktheHumanGeneticsSampleBankforprocessingofsamples.
OSUColumbusareacontrolspecimenswereprovidedbytheOhioStateUniversity'sHumanGeneticsSampleBank.
ThePBCSthanksMarkSherman,NeonilaSzeszenia-Dabrowska,BeataPeplonska,WitoldZatonski,PeiChaoandMichaelStagner.
TheRBCSthanksPetraBos,JannetBlom,EllenCrepin,AnjaNieuwlaat,AnnetteHeemskerkandtheErasmusMCFamilyCancerClinic.
TheSBCSthanksSueHigham,IanBrock,SabapathyBalasu-bramanian,HelenCrampandDanConnley.
TheSEARCHthankstheSEARCHandEPIC-Norfolkteams.
TheiCOGSstudywouldnothavebeenpossiblewithoutthecontributionsofthefollowing:QinWang(BCAC),AndrewBerchuck(OCAC),RosalindA.
Eeles,AliAminAlOlama,ZsofiaKote-Jarai,SaraBenlloch(PRACTICAL),AntonisAntoniou,LesleyMcGuffogandKenOffit(CIMBA),AndrewLee,andEdDicks,CraigLuccariniandthestaffoftheCentreforGeneticEpidemiologyLaboratory,AnnaGonzalez-NeiraandthestaffoftheCNIOgenotypingunit,DanielC.
Tessier,FrancoisBacot,DanielVincent,SylvieLaBoissie`reandFredericRobidouxandthestaffoftheMcGillUniversityandGenomeQuebecInnovationCentre,andthestaffoftheCopenhagenDNAlaboratory,andJulieM.
Cunningham,SharonA.
Windebank,ChristopherA.
Hilker,JeffreyMeyerandthestaffofMayoClinicGenotypingCoreFacility.
ConsortiamembersGENICANetwork.
HiltrudBrauch,Wing-YeeLo,ChristinaJustenhoven:Dr.
MargareteFischer-Bosch-InstituteofClinicalPharma-cology,Stuttgart,andUniversityofTu¨bingen,Germany.
Yon-DschunKo,ChristianBaisch:DepartmentofInternalMedicine,EvangelischeKlinikenBonngGmbH,JohanniterKrankenhaus,Bonn,Germany.
Hans-PeterFischer:InstituteofPathology,UniversityofBonn,Bonn,Germany.
UteHamann:MolecularGeneticsofBreastCancer,DeutschesKrebs-forschungszentrum(DKFZ),Heidelberg,Germany.
ThomasBru¨ning,BeatePesch,SylviaRabstein,AnneLotz:InstituteoftheRuhrUniversityBochum(IPA),Bochum,Germany.
VolkerHarth:InstituteforOccupa-tionalMedicineandMaritimeMedicine,UniversityMedicalCenterHamburg-Eppendorf,Germany.
kConFabInvestigators.
Seehttp://www.
kconfab.
org/Organisation/Members.
aspxAOCS.
Seehttp://www.
aocstudy.
org/org_coll.
aspAuthorContributionsConceivedanddesignedtheexperiments:HNDGGCTACRLMDFESKKMJCCADMSMGCPH.
Performedtheexperiments:SKDGKMRLMDFE.
Analyzedthedata:SKDGKMRLMHNDFE.
Contributedreagents/materials/analysistools:SKHNDGKMGCTACRLMPDPPUHMKSA.
MeindlRWTHCBK.
AaltonenGGGDFEPAFMJHILAH.
BrauchQWEJSH.
BrennerAKDMSGFLTAMK.
Aittoma¨kiJ.
LiuPHAIKHJ.
LiKCJCCRHARPSDFJOFJPIdSSNJLGZAJLHHTM.
BuiEMDFSMCSCAJ.
StoneHMHMAARBvdLA.
MannermaaRKSBMMPLCTNRSJCDJHSSCMWRRABLJVVFBHMGSABEMWBSEBBGNSFNHFPMZJIAPJ.
BenitezCAHBEHFSLLMAMDMSRLJ.
BrownFJCXWCVJEODLMMRPMRCPGTTPLPC.
MulotFMA.
SchneeweissC.
SohnBBITMJKNMJAKSTAMMNVBNNATDHACHDMEMGCJFJ.
LissowskaLBPDRAEMTC.
SeynaeveCJvAVNKSSAETCBADYALSMPRPPM.
BarilePMJWMMJMCA.
JagerA.
JakubowskaJ.
LubinskiKJBKDC.
McLeanTBYDKVAC.
StegmaierA.
SwerdlowAANOMJJ.
SimardMDKPAJVMGVKMKBJDVMKJMHkConFabInvestigatorsAustralianOvarianCancerStudyGroupTheGENICANetwork.
Wrotethepaper:SKHNRLMAC.
Providedcriticalreviewofthemanuscript:SKHNDGKMGCTACRLMPDPPUHMKSA.
MeindlRWTHCBK.
AaltonenGGGDFEPAFMJHILAH.
BrauchQWEJSH.
BrennerAKDMSGFLTAMK.
Aittoma¨kiJ.
LiuPHAIKHJ.
LiKCJCCRHARPSDFJOFJPIdSSNJLGZAJLHHTM.
BuiEMDFSMCSCAJ.
StoneHMHMAARBvdLA.
MannermaaRKSBMMPLCTNRSJCDJHSSCMWRRABLJVVFBHMGSABEMWBSEBBGNSFNHFPMZJIAPJ.
BenitezCAHBEHFSLLMAMDMSRLJ.
BrownFJCXWCVJEODLMMRPMRCPGTTPLPC.
MulotFMA.
SchneeweissC.
SohnBBITMJKNMJAKSTAMMNVBNNATDHACHDMEMGCJFJ.
LissowskaLBPDRAEMTC.
SeynaeveCJvAVNKSSAETCBADYALPLOSONE|www.
plosone.
org10November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRiskSMPRPPM.
BarilePMJWMMJMCA.
JagerA.
JakubowskaJ.
LubinskiKJBKDC.
McLeanTBYDKVAC.
StegmaierA.
SwerdlowAANOMJJ.
SimardMDKPAJVMGVKMKBJDVMKJMHkConFabInvestigatorsAustralianOvarianCancerStudyGroupTheGENICANetwork.
Approvedthefinalversionofthemanuscript:SKHNDGKMGCTACRLMPDPPUHMKSA.
MeindlRWTHCBK.
AaltonenGGGDFEPAFMJHILAH.
BrauchQWEJSH.
BrennerAKDMSGFLTAMK.
Aittoma¨kiJ.
LiuPHAIKHJ.
LiKCJCCRHARPSDFJOFJPIdSSNJLGZAJLHHTM.
BuiEMDFSMCSCAJ.
StoneHMHMAARBvdLA.
MannermaaRKSBMMPLCTNRSJCDJHSSCMWRRABLJVVFBHMGSABEMWBSEBBGNSFNHFPMZJIAPJ.
BenitezCAHBEHFSLLMAMDMSRLJ.
BrownFJCXWCVJEODLMMRPMRCPGTTPLPC.
MulotFMA.
SchneeweissC.
SohnBBITMJKNMJAKSTAMMNVBNNATDHACHDMEMGCJFJ.
LissowskaLBPDRAEMTC.
SeynaeveCJvAVNKSSAETCBADYALSMPRPPM.
BarilePMJWMMJMCA.
JagerA.
JakubowskaJ.
LubinskiKJBKDC.
McLeanTBYDKVAC.
StegmaierA.
SwerdlowAANOMJJ.
SimardMDKPAJVMGVKMKBJDVMKJMHkConFabInvestigatorsAustralianOvarianCancerStudyGroupTheGENICANetwork.
Administrativetechnicalormaterialsupport:MKBJDMSRL.
References1.
JemalA,BrayF,CenterMM,FerlayJ,WardE,etal.
(2011)Globalcancerstatistics.
CACancerJClin61:69–90.
2.
LichtensteinP,HolmNV,VerkasaloPK,IliadouA,KaprioJ,etal.
(2000)Environmentalandheritablefactorsinthecausationofcancer—analysesofcohortsoftwinsfromSweden,Denmark,andFinland.
NEnglJMed343:78–85.
3.
GhoussainiM,FletcherO,MichailidouK,TurnbullC,SchmidtMK,etal.
(2012)Genome-wideassociationanalysisidentifiesthreenewbreastcancersusceptibilityloci.
NatGenet44:312–318.
4.
MichailidouK,HallP,Gonzalez-NeiraA,GhoussainiM,DennisJ,etal.
(2013)Large-scalegenotypingidentifies41newlociassociatedwithbreastcancerrisk.
NatGenet45:353–361,361e351–352.
5.
DenliAM,TopsBB,PlasterkRH,KettingRF,HannonGJ(2004)ProcessingofprimarymicroRNAsbytheMicroprocessorcomplex.
Nature432:231–235.
6.
LeeY,AhnC,HanJ,ChoiH,KimJ,etal.
(2003)ThenuclearRNaseIIIDroshainitiatesmicroRNAprocessing.
Nature425:415–419.
7.
HutvagnerG,McLachlanJ,PasquinelliAE,BalintE,TuschlT,etal.
(2001)AcellularfunctionfortheRNA-interferenceenzymeDicerinthematurationofthelet-7smalltemporalRNA.
Science293:834–838.
8.
SosioM,KloostermanH,BianchiA,deVreugdP,DijkhuizenL,etal.
(2004)OrganizationoftheteicoplaningeneclusterinActinoplanesteichomyceticus.
Microbiology150:95–102.
9.
FilipowiczW,BhattacharyyaSN,SonenbergN(2008)Mechanismsofpost-transcriptionalregulationbymicroRNAs:aretheanswersinsightNatRevGenet9:102–114.
10.
ShuklaGC,SinghJ,BarikS(2011)MicroRNAs:Processing,Maturation,TargetRecognitionandRegulatoryFunctions.
MolCellPharmacol3:83–92.
11.
KrolJ,LoedigeI,FilipowiczW(2010)ThewidespreadregulationofmicroRNAbiogenesis,functionanddecay.
NatRevGenet11:597–610.
12.
ZhongX,CoukosG,ZhangL(2012)miRNAsinhumancancer.
MethodsMolBiol822:295–306.
13.
FriedmanRC,FarhKK,BurgeCB,BartelDP(2009)MostmammalianmRNAsareconservedtargetsofmicroRNAs.
GenomeRes19:92–105.
14.
FaraziTA,HoellJI,MorozovP,TuschlT(2013)MicroRNAsinHumanCancer.
AdvExpMedBiol774:1–20.
15.
WangX,ChenX,WangR,XiaoP,XuZ,etal.
(2013)microRNA-200cmodulatestheepithelial-to-mesenchymaltransitioninhumanrenalcellcarcinomametastasis.
OncolRep30:643–650.
16.
LiangZ,WuH,XiaJ,LiY,ZhangY,etal.
(2010)InvolvementofmiR-326inchemotherapyresistanceofbreastcancerthroughmodulatingexpressionofmultidrugresistance-associatedprotein1.
BiochemPharmacol79:817–824.
17.
VoliniaS,CroceCM(2013)PrognosticmicroRNA/mRNAsignaturefromtheintegratedanalysisofpatientswithinvasivebreastcancer.
ProcNatlAcadSciUSA110:7413–7417.
18.
GuoL,ZhaoY,YangS,CaiM,WuQ,etal.
(2012)Genome-widescreenforaberrantlyexpressedmiRNAsrevealsmiRNAprofilesignatureinbreastcancer.
MolBiolRep.
19.
(2012)Comprehensivemolecularportraitsofhumanbreasttumours.
Nature490:61–70.
20.
LandiD,GemignaniF,NaccaratiA,PardiniB,VodickaP,etal.
(2008)Polymorphismswithinmicro-RNA-bindingsitesandriskofsporadiccolorectalcancer.
Carcinogenesis29:579–584.
21.
SongF,ZhengH,LiuB,WeiS,DaiH,etal.
(2009)AnmiR-502-bindingsitesingle-nucleotidepolymorphisminthe3'-untranslatedregionoftheSET8geneisassociatedwithearlyageofbreastcanceronset.
ClinCancerRes15:6292–6300.
22.
KontorovichT,LevyA,KorostishevskyM,NirU,FriedmanE(2010)SinglenucleotidepolymorphismsinmiRNAbindingsitesandmiRNAgenesasbreast/ovariancancerriskmodifiersinJewishhigh-riskwomen.
IntJCancer127:589–597.
23.
NicolosoMS,SunH,SpizzoR,KimH,WickramasingheP,etal.
(2010)Single-nucleotidepolymorphismsinsidemicroRNAtargetsitesinfluencetumorsusceptibility.
CancerRes70:2789–2798.
24.
HiardS,CharlierC,CoppietersW,GeorgesM,BaurainD(2010)Patrocles:adatabaseofpolymorphicmiRNA-mediatedgeneregulationinvertebrates.
NucleicAcidsRes38:D640–651.
25.
DiteGS,JenkinsMA,SoutheyMC,HockingJS,GilesGG,etal.
(2003)Familialrisks,early-onsetbreastcancer,andBRCA1andBRCA2germlinemutations.
JNatlCancerInst95:448–457.
26.
FletcherO,JohnsonN,PallesC,dosSantosSilvaI,McCormackV,etal.
(2006)InconsistentassociationbetweentheSTK15F31Igeneticpolymorphismandbreastcancerrisk.
JNatlCancerInst98:1014–1018.
27.
HunterDJ,KraftP,JacobsKB,CoxDG,YeagerM,etal.
(2007)Agenome-wideassociationstudyidentifiesallelesinFGFR2associatedwithriskofsporadicpostmenopausalbreastcancer.
NatGenet39:870–874.
28.
FrankB,HemminkiK,WappenschmidtB,MeindlA,KlaesR,etal.
(2006)AssociationoftheCASP10V410IvariantwithreducedfamilialbreastcancerriskandinteractionwiththeCASP8D302Hvariant.
Carcinogenesis27:606–609.
29.
Flesch-JanysD,SlangerT,MutschelknaussE,KroppS,ObiN,etal.
(2008)Riskofdifferenthistologicaltypesofpostmenopausalbreastcancerbytypeandregimenofmenopausalhormonetherapy.
IntJCancer123:933–941.
30.
LiJ,HumphreysK,HeikkinenT,AittomakiK,BlomqvistC,etal.
(2011)Acombinedanalysisofgenome-wideassociationstudiesinbreastcancer.
BreastCancerResTreat126:717–727.
31.
LeuM,HumphreysK,SurakkaI,RehnbergE,MuiluJ,etal.
(2010)NordicDB:aNordicpoolandportalforgenome-widecontroldata.
EurJHumGenet18:1322–1326.
32.
TurnbullC,AhmedS,MorrisonJ,PernetD,RenwickA,etal.
(2010)Genome-wideassociationstudyidentifiesfivenewbreastcancersusceptibilityloci.
NatGenet42:504–507.
33.
EastonDF,PooleyKA,DunningAM,PharoahPD,ThompsonD,etal.
(2007)Genome-wideassociationstudyidentifiesnovelbreastcancersusceptibilityloci.
Nature447:1087–1093.
34.
LiangD,MeyerL,ChangDW,LinJ,PuX,etal.
(2010)GeneticvariantsinMicroRNAbiosynthesispathwaysandbindingsitesmodifyovariancancerrisk,survival,andtreatmentresponse.
CancerRes70:9765–9776.
35.
Ruiz-RuizC,Munoz-PinedoC,Lopez-RivasA(2000)Interferon-gammatreatmentelevatescaspase-8expressionandsensitizeshumanbreasttumorcellstoadeathreceptor-inducedmitochondria-operatedapoptoticprogram.
CancerRes60:5673–5680.
36.
BarrettJH,IlesMM,HarlandM,TaylorJC,AitkenJF,etal.
(2011)Genome-wideassociationstudyidentifiesthreenewmelanomasusceptibilityloci.
NatGenet43:1108–1113.
37.
deMartinoM,HaitelA,SchatzlG,KlinglerHC,KlatteT(2013)TheCASP8-6526NInsertion/DeletionPromoterPolymorphismIsAssociatedwithRenalCellCarcinomaRiskandMetastasis.
JUrol.
38.
CoxA,DunningAM,Garcia-ClosasM,BalasubramanianS,ReedMW,etal.
(2007)AcommoncodingvariantinCASP8isassociatedwithbreastcancerrisk.
NatGenet39:352–358.
39.
PengS,LuB,RuanW,ZhuY,ShengH,etal.
(2011)Geneticpolymorphismsandbreastcancerrisk:evidencefrommeta-analyses,pooledanalyses,andgenome-wideassociationstudies.
BreastCancerResTreat127:309–324.
40.
MacPhersonG,HealeyCS,TeareMD,BalasubramanianSP,ReedMW,etal.
(2004)AssociationofacommonvariantoftheCASP8genewithreducedriskofbreastcancer.
JNatlCancerInst96:1866–1869.
41.
FrankB,BermejoJL,HemminkiK,KlaesR,BugertP,etal.
(2005)Re:AssociationofacommonvariantoftheCASP8genewithreducedriskofbreastcancer.
JNatlCancerInst97:1012;authorreply1012–1013.
42.
LinWY,CampNJ,GhoussainiM,BeesleyJ,MichailidouK,etal.
(2014)IdentificationandcharacterizationofnovelassociationsintheCASP8/ALS2CR12regiononchromosome2withbreastcancerrisk.
HumMolGenet.
43.
SunD,LeeG,LeeJH,KimHY,RheeHW,etal.
(2010)AmetazoanorthologofSpoThydrolyzesppGppandfunctionsinstarvationresponses.
NatStructMolBiol17:1188–1194.
44.
KilpinenS,AutioR,OjalaK,IljinK,BucherE,etal.
(2008)Systematicbioinformaticanalysisofexpressionlevelsof17,330humangenesacross9,783samplesfrom175typesofhealthyandpathologicaltissues.
GenomeBiol9:R139.
45.
LewisBP,BurgeCB,BartelDP(2005)Conservedseedpairing,oftenflankedbyadenosines,indicatesthatthousandsofhumangenesaremicroRNAtargets.
Cell120:15–20.
46.
KerteszM,IovinoN,UnnerstallU,GaulU,SegalE(2007)TheroleofsiteaccessibilityinmicroRNAtargetrecognition.
NatGenet39:1278–1284.
47.
SungH,LeeKM,ChoiJY,HanS,LeeJY,etal.
(2011)CommongeneticpolymorphismsofmicroRNAbiogenesispathwaygenesandriskofbreastcancer:acase-controlstudyinKorea.
BreastCancerResTreat130:939–951.
PLOSONE|www.
plosone.
org11November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRisk48.
BirrerMJ,SegalS,DeGreveJS,KayeF,SausvilleEA,etal.
(1988)L-myccooperateswithrastotransformprimaryratembryofibroblasts.
MolCellBiol8:2668–2673.
49.
WuR,LinL,BeerDG,EllensonLH,LambBJ,etal.
(2003)AmplificationandoverexpressionoftheL-MYCproto-oncogeneinovariancarcinomas.
AmJPathol162:1603–1610.
50.
XiongF,WuC,ChangJ,YuD,XuB,etal.
(2011)GeneticvariationinanmiRNA-1827bindingsiteinMYCL1alterssusceptibilitytosmall-celllungcancer.
CancerRes71:5175–5181.
51.
LombardoF,KomatsuD,HadjiargyrouM(2004)MolecularcloningandcharacterizationofMustang,anovelnuclearproteinexpressedduringskeletaldevelopmentandregeneration.
FASEBJ18:52–61.
52.
KapusheskyM,AdamusiakT,BurdettT,CulhaneA,FarneA,etal.
(2012)GeneExpressionAtlasupdate—avalue-addeddatabaseofmicroarrayandsequencing-basedfunctionalgenomicsexperiments.
NucleicAcidsRes40:D1077–1081.
53.
WitkosTM,KoscianskaE,KrzyzosiakWJ(2011)PracticalAspectsofmicroRNATargetPrediction.
CurrMolMed11:93–109.
PLOSONE|www.
plosone.
org12November2014|Volume9|Issue11|e109973MicroRNARelatedPolymorphismsandBreastCancerRisk
- Liguejusewang相关文档
- Subjectjusewang
- examinedjusewang
- attractjusewang
- Alcjusewang
- Seriesjusewang
- selectionjusewang
PacificRack 下架旧款方案 续费涨价 谨慎自动续费
前几天看到网友反馈到PacificRack商家关于处理问题的工单速度慢,于是也有后台提交个工单问问,没有得到答复导致工单自动停止,不清楚商家最近在调整什么。而且看到有网友反馈到,PacificRack 商家的之前年付低价套餐全部下架,而且如果到期续费的话账单中的产品价格会涨价不少。所以,如果我们有需要续费产品的话,谨慎选择。1、特价产品下架我们看到他们的所有原来发布的特价方案均已下架。如果我们已有...
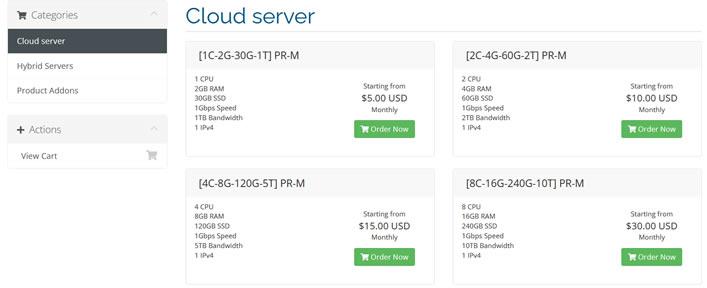
Megalayer(月599元)限时8月香港和美国大带宽服务器
第一、香港服务器机房这里我们可以看到有提供四个大带宽方案,是全向带宽和国际带宽,前者适合除了中国大陆地区的全网地区用户可以用,后者国际带宽适合欧美地区业务。如果我们是需要大陆地区速度CN2优化的,那就需要选择常规的优化带宽方案,参考这里。CPU内存硬盘带宽流量价格选择E3-12308GB240GB SSD50M全向带宽不限999元/月方案选择E3-12308GB240GB SSD100M国际带宽不...

GreenCloudVPS($30/年),500G大硬盘VPS,10Gbps带宽
GreenCloudVPS最近在新加坡DC2节点上了新机器,Dual Xeon Silver 4216 CPU,DDR4内存,10Gbps网络端口,推出了几款大硬盘VPS套餐,基于KVM架构,500GB磁盘起年付30美元。除了大硬盘套餐外,还加推了几款采用NVMe硬盘的常规套餐,最低年付20美元。不过需要提醒的是,机房非直连中国,尤其是电信用户ping值感人,包括新加坡DC1也是如此。大硬盘VPS...
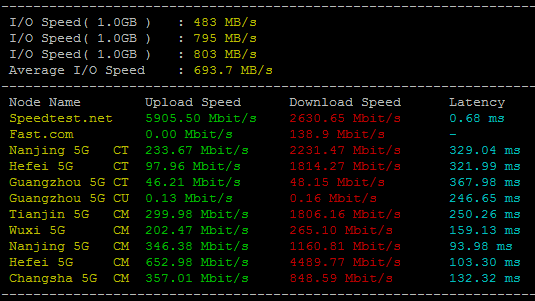
jusewang为你推荐
-
ovOV摄像头是哪个国家的安装程序配置服务器失败安装用友T3出现安装程序配置服务器失败是怎么回事公章制作如何制作公章微信如何建群微信怎么建立群今日热点怎么删除如何彻底删除今日热点网站推广外链网站推广,免费的超级外链有用吗?seo还应该做什么网站推广外链在网站推广中,有着一种“购买外链”是什么意思去鼠标加速度怎样去除电脑鼠标加速?手工杀毒怎样不用杀毒软件自己手动查毒?msn与qqMSN与QQ的区别?