activationcjblaze
cjblaze 时间:2021-01-14 阅读:()
1Runningtitle:VQ29regulatesseedlingde-etiolationResearcharea:SignalingandResponseCorrespondingauthor:RongchengLin,86-10-62836905,e-mail:rclin@ibcas.
ac.
cnKeyLaboratoryofPhotobiology,InstituteofBotany,ChineseAcademyofSciences,Beijing100093,ChinaOne-sentencesummary:AnewtranscriptionregulatorVQ29interactswiththetranscriptionfactorPIF1tomodulatehypocotylcellgrowthinresponsetolight.
PlantPhysiologyPreview.
PublishedonFebruary25,2014,asDOI:10.
1104/pp.
113.
234492Copyright2014bytheAmericanSocietyofPlantBiologistshttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
2ArabidopsisVQ-motif-containingProtein29RepressesSeedlingDe-etiolationbyInteractingwithPIF1YunliangLi,a,bYanjunJing,aJunjiaoLi,a,cGangXu,a,bRongchengLina,1aKeyLaboratoryofPhotobiology,InstituteofBotany,theChineseAcademyofSciences,Beijing100093,ChinabUniversityoftheChineseAcademyofSciences,Beijing100049,ChinacCollegeofLifeSciences,CapitalNormalUniversity,Beijing100048,China1Correspondingauthor,e-mail:rclin@ibcas.
ac.
cnTheauthorresponsiblefordistributionofmaterialintegraltothefindingspresentedinthisarticleinaccordancewiththepolicydescribedintheinstructionsforauthors(www.
plantphysiol.
org)isRongchengLin(rclin@ibcas.
ac.
cn).
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
3Footnotes:ThisworkwassupportedbygrantsfromtheNationalNaturalScienceFoundationofChina(31170221,31325002),andtheMinistryofAgricultureofChina(2011ZX08009-003)toR.
L.
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
4ABSTRACTSeedlingde-etiolation,acriticalprocessinearlyplantdevelopment,isregulatedbyanintricatetranscriptionalnetwork.
Here,weidentifiedVQmotif-containingprotein29(VQ29)asanovelregulatorofthephotomorphogenicresponseinArabidopsisthaliana.
Weshowedthat29ofthe34VQproteinspresentinArabidopsisexhibittranscriptionalactivityinplantcells,andthatmutationsintheVQmotifaffectthetranscriptionalactivityofVQ29.
WethenfunctionallycharacterizedVQ29andshowedthatthehypocotylgrowthofplantsoverexpressingVQ29ishyposensitivetofar-redandlowintensitywhitelight,whereasavq29loss-of-functionmutantexhibitsdecreasedhypocotylelongationunderlowintensityoffar-redlightorwhitelight.
Consistentwiththis,VQ29expressionisrepressedbylightinaphytochrome-dependentmanner.
Intriguingly,ouryeasttwo-hybrid,bimolecularfluorescencecomplementationandco-immunoprecipitationassaysshowedthatVQ29physicallyinteractswithPHYTOCHROME-INTERACTINGFACTOR1(PIF1).
WethenshowedthatVQ29andPIF1directlybindtothepromoterofacellelongation-relatedgene,XYLOGLUCANENDOTRANSGLYCOSYLASE7,andco-activateitsexpression.
Furthermore,thevq29pif1doublemutanthasshorterhypocotylsthaneitherofthecorrespondingsinglemutants.
Therefore,ourstudyrevealsthatVQ29isanegativetranscriptionalregulatoroflight-mediatedinhibitionofhypocotylelongationthatlikelypromotesthetranscriptionalactivityofPIF1duringearlyseedlingdevelopment.
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
5INTRODUCTIONLightisanimportantenvironmentalsignalthataffectsplantgrowthanddevelopmentthroughoutitslifecycle,directingprocessessuchasseedgermination,seedlingde-etiolation,phototropism,circadianrhythms,shadeavoidance,andfloweringtiming.
Dark-grownseedlings,whichadoptadevelopmentalprogramknownasetiolationorskotomorphogenesis,exhibitelongatedhypocotylsandclosedcotyledonswithapicalhooks.
Uponlightirradiation,seedlingsundergode-etiolationorphotomorphogenesis,whichslowshypocotylgrowthandcausescotyledonstoexpandandchloroplastsandchlorophyllstodevelop(vonArnimandDeng,1996).
Intensiveresearchhasrevealedthemainsignalingpathwaygoverningphotomorphogenesis(Chenetal.
,2004,LauandDeng,2010,Arsovskietal.
,2012).
Toinitiatethelightresponses,plantsrelyonasetofphotoreceptors,includingthered/far-redlight-absorbingphytochromes(phys)andtheblue/UV-Alight-absorbingcryptochromes(crys).
Activationofphotoreceptorstransmitssignalstokeydownstreamnegativefactors,suchasCOP1(CONSTITUTIVEPHOTOMORPHOGENIC1)andmembersofthePIF(PHYTOCHROMEINTERACTINGFACTOR)proteinfamily.
COP1isaRING-typeE3ubiquitinligasethattargetsphotomorphogenesis-promotingfactors,suchasHY5(ELONGATEDHYPOCOTYL5)andHFR1(LONGHYPOCOTYLINFAR-RED1),for26Sproteasome-mediateddegradation,whichdesensitizesthelightpathwayinitiatedbybothphysandcrys(WeiandDeng,1996,LauandDeng,2012).
PIFsencodeagroupofbasichelix-loop-helix(bHLH)transcriptionfactors(TFs)thatarephosphorylatedanddegradedinaphy-dependentmannerinlight.
PIFproteins(includingPIF1,3,4,and5)playredundantrolesindirectlyregulatinggeneexpressionandrepressingphotomorphogenicresponses(Leivaretal.
,2008,Shinetal.
,2009,LeivarandQuail,2010).
GeneticandmolecularstudiesinArabidopsisthalianahaveidentifiedanotherseriesofsignalingcomponentsthatisinvolvedineithernegativelyorpositivelyregulatingthelightpathway(Jiaoetal.
,2007).
Notably,manyoftheidentifiedhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
6effectorsarenuclear-localizedTFs,suchasFHY3andFAR1(transposase-derivedTFs),bZIP16(bZIPTFs),HFR1andMYC2(bHLHTFs),LAF1(aMYBTF),andSTH2andLZF1(B-box-containingTFs)(Hudsonetal.
,1999,WangandDeng,2002,Ballesterosetal.
,2001,Hsiehetal.
,2012,Dattaetal.
,2007,Changetal.
,2011,Yadavetal.
,2005).
Moreover,genesencodingF-boxproteins(suchasEID1andAFR),kinases(e.
g.
,NDPK2),andphosphatases(suchasPP7)werealsoisolatedandshowntomediatelightsignaling(Dieterleetal.
,2001,HarmonandKay,2003,Choietal.
,1999,Mlleretal.
,2003).
However,theregulatorymechanismsunderlyingtheactionsofthesefactorsarenotwellunderstood.
Inaddition,werecentlyshowedthatachromatinremodelingfactor,EPP1(enhancedphotomorphogenesis1)/PICKLE,interactswithHY5tofine-tunelightsignalingbymodulatingH3k27me3levelsoncellelongation-relatedgenes(Jingetal.
,2013).
Eventhoughawiderangeoflightsignalingintermediateshavebeenidentifiedandextensivelystudied,acompletepictureofthelightsignalingpathwayhasyettoemerge.
AgroupofgenesencodingproteinscontainingauniqueandconservedFxxxVQxxTGmotif(termedtheVQmotif)wasrecentlyidentified(Xieetal.
,2010,Chengetal.
,2012).
Theseproteinsinclude34membersinArabidopsisandaredesignatedasVQmotif-containingproteins(VQ).
Thefunctionofafewmembersofthisfamilyhasbeencharacterizedtodate.
Forexample,sigmafactorbindingprotein1(SIB1/VQ23),itsclosehomologSIB2(VQ16),andMAPkinase4substrate1(MKS1/VQ21)arerequiredfortheplantdefenseresponse(Narusakaetal.
,2008,Xieetal.
,2010,Laietal.
,2011,Andereassonetal.
,2005).
Inaddition,IKU1(VQ14)isaregulatorofendospermgrowthandseedsize(Wangetal.
,2010),whileCaMBP25(VQ15)andVQ9functionasnegativeeffectorsofosmoticandsalinitystresstolerance,respectively(Perrucetal.
,2004,Huetal.
,2013).
Itisanticipatedthatthisfamilyrespondstovariousenvironmentalsignalsandplaysdiverserolesinplantdefense,growth,anddevelopment.
However,thefunctionsandregulatorymechanismsofmostVQfamilymembersremainunknown.
Inthisstudy,wedemonstratethattheVQfamilyofproteinslargelypossessestranscriptionalactivities.
Furthermore,wecharacterizedtheregulationandfunctionofhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
7VQ29indetail.
WeshowthatVQ29expressionisdown-regulatedbylight.
OverexpressionofVQ29resultsinhyposensitivityofhypocotylgrowthtofar-redandlowlightconditions,whereasthevq29loss-of-functionmutantexhibitsdecreasedhypocotylelongationunderlowintensityoffar-redandwhitelight,duringseedlingde-etiolation.
WealsodemonstratethatVQ29physicallyinteractswithPIF1andthattheseproteinscooperativelyactivatetheexpressionofdownstreamgenes.
OurstudyidentifiesanovelfactorinphotomorphogenesisandprovidesinsightintotherolesofVQfamilyproteinsinregulatingdiverseplantgrowthanddevelopmentalprocesses.
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
8RESULTSAnalysisofVQgenesfromArabidopsis,rice,andmossPreviousstudiesdocumentedthattheVQgenefamilyisfoundonlyinplantsanditwassystematicallystudiedinArabidopsisthaliana(Xieetal.
,2010,Chengetal.
,2012).
Togaininsightintotheevolutionofthisfamily,wesearchedtheGenBankdatabaseforsequencesoftheVQgenesfromrice(Oryzasativa)andmoss(Physcomitrellapatens),whichrepresentmonocotandlowerplants,respectively.
WhereasthemodeldicotplantArabidopsishas34VQmembers,riceandmosshave39and25,respectively.
Interestingly,ArabidopsisandriceVQproteinsarerelativelysmall,withabout85%and92%oftheproteinscontainingfewerthan300aminoacidresidues,respectively.
Incontrast,68%ofthemossVQproteinsarelongerthan300aminoacids(SupplementalFigureS1A).
WefurtherfoundthatmostVQgenesinhigherplants(30inArabidopsisand37inrice)donotcontainanintron.
However,inmoss,only7VQgenesdonothaveintron,whereas5VQshaveoneintronand13genespossesstwoormoreintrons(SupplementalFigureS1B).
TheseresultssuggestthatVQgenestendtobeintronlessandencoderelativelysmallproteinsinhigherplants.
VQproteinsexhibittranscriptionalactivityToinvestigatewhethertheVQproteinsareinvolvedintranscriptionalregulation,weattemptedtoisolatetheopenreadingframe(ORF)ofeachVQgeneinArabidopsisColumbia(Col)plantsusingreversetranscriptionpolymerasechainreaction(RT-PCR,forVQ2)orPCRforotherintronlessVQgenes.
ThePCRprimersweredesignedaccordingtotheavailablecDNAsequenceinformationorthepredictedsequences.
TheORFsofallVQgeneswereamplifiedandclonedintothepEASYvectorandverifiedbysequencing.
WethensubclonedtheVQgenesin-framewiththeGAL4DNA-bindingdomain(GBD)andunderthecontrolofthecauliflowermosaicvirus(CaMV)35SpromoterinthepSAT-GAL4DBvector(Jingetal.
,2013).
Theconstructwasco-transformedwithaluciferasereportergene(LUC),drivenbythe35Sminimalhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
9promoterandfusedin-frametoaGAL4bindingsequence,intoArabidopsismesophyllprotoplasts(Figure1A).
WefoundthatGBDfusionproteinswithVQ14,VQ5,VQ15,VQ16,VQ9,VQ23,VQ3,VQ24,VQ34,VQ17,VQ32,orVQ30drasticallyactivatedLUCreporterexpressioncomparedwithGBDalone.
TheVQ26,VQ12,VQ18,VQ28,andVQ6fusionspromotedLUCexpressiontoalesserextent(Figure1B).
However,GBDfusionswithVQ19,VQ31,VQ4,VQ8,VQ13,VQ33,VQ11,VQ29,VQ7,VQ2,VQ21,orVQ20,remarkablyrepressedtheexpressionoftheLUCreporter(Figure1C).
WealsoobservedthatGBDfusionswithVQ22,VQ1,VQ27,VQ25,andVQ10didnotaffectthetranscriptionofLUC(Figure1B,C).
ThesedataindicatethatmostmembersoftheVQfamilypossesseithertranscriptionalactivationorrepressionactivitiesinthisheterologousgenereportersystem.
Inthisstudy,wefocusedonVQ29(At4g37710)becauseitisinvolvedinseedlingde-etiolationresponse(seebelowindetail).
MutationintheVQmotifaffectstranscriptionalactivitySinceVQfamilymemberscontainonlytheVQmotifandpossessactivationorrepressionactivity,weaskedwhethertheVQmotifisrequiredfortranscriptionalactivity.
WethenalteredconservedaminoacidsintheVQmotifofVQ29,wheretheveryhydrophobicresiduevaline(V)waschangedintoalesshydrophobicresiduealanine(A)orthehydrophilicresidueasparticacid(D),andthehydrophilicresidueglutamine(Q)wasmutagenizedintothehydrophobicresidueleucine(L).
AsshowninFigure1D,singlemutationofVQ29(V70A)orVQ29(V70D)abolishedtherepressiveactivityofVQ29,andgreatlyactivatedLUCreportergeneexpression,whereasmutationinVQ29(Q71L)didnotaffecttheactivity.
Furthermore,doublemutationsinVQ29(V70D,Q71L)ledtoasignificantinductionofLUCtoalesserextentthanVQ29(V70D)(Figure1D).
Itshouldbenotedthattwopointmutations(V70DandQ71L)astested,didnotaffectthelevelsoftheVQ29protein(SupplementalFigureS2).
Takentogether,theseresultssuggestthattheVQmotifislikelyinvolvedinmediatingthetranscriptionalactivityofVQproteins.
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
10OverexpressionofVQ29reducesthehypocotylgrowthresponseunderfar-redandlowintensityofwhitelightconditionsToelucidatethebiologicalfunctionofVQ29,weobtainedtwoT-DNAinsertionlinesofVQ29,Salk_061586andSalk_061438.
PCRgenotypingandsequencingstudiesrevealedthattheT-DNAisinsertedinthepromoterregion136basepairsupstreamoftheATGstartcodonofVQ29inbothmutants.
Thesemutantsthusrepresentthesameallele(Figure2A).
TheyhadbeenbackcrossedfivetimeswiththeColwildtypeandthusthepotentialbackgroundmutationswerelargelyeliminated.
QuantitativeRT-PCRanalysisshowedthattheVQ29transcriptswerebarelydetectableinthemutant(hereafterreferredtoasvq29-1),suggestingthatitisanullallele(Figure2B).
Wealsogeneratedtransgenicplantsover-expressingVQ29,fusedwithaMYCtaganddrivenbytheCaMV35Spromoter(Pro35S:Myc-VQ29,VQ29-OE).
Overfortytransgeniclineswereobtained,twoofwhich(lines#10and#14)werefurtherstudiedinthefollowingexperiments.
ImmunoblottinganalysisusingtheMYCantibodyshowedthatbothlinesaccumulatedMyc-VQ29fusionprotein,withline#10havingahigherlevelofexpressionthanline#14(Figure2C).
Wethentestedthehypocotylgrowthresponseofthevq29-1mutantandVQ29overexpressionlinesundervariouslightconditions.
Thehypocotyllengthofvq29-1wasslightlybutsignificantlyshorterthanthewildtypeunderlowintensityofwhitelight(lessthan20molm-2s-1)orlowintensityoffar-redlight(lessthan12molm-2s-1),butwasindistinguishablefromthatofthewildtypeinredorbluelightconditionswithmultiplefluenceratestested(Figure2D,2E,SupplementalFigureS3).
However,thehypocotylsofVQ29-OEtransgenicseedlingsweresignificantlylongerthanthoseofwild-typeplantsunderfar-redconditions,withline#10exhibitingthestrongerphenotype,consistentwithitshigherVQ29level(Figure2C-E,SupplementalFigureS3).
Thehypocotylsofoverexpressionline#10werealsolongerthanthewildtypeunderlowwhitelightconditions,butnotindarknessorinredorbluelightconditions(Figure2E,SupplementalFigureS3).
Ithasbeendocumentedthatsucrosepromotesseedlinggrowth(Stewartetal.
,2011).
EvenwithoutsucrosesupplementintheMSmedia,vq29-1exhibitedreducedhypocotylelongationunderlowwhitelightandhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
11differentintensitiesoffar-redlight,andthehypocotyllengthofVQ29-OEplants(line#10)waslongerthanthewildtypeundertheseconditions(SupplementalFigureS4).
Moreover,theexpressionoftwolight-responsivegenesinvolvedincellelongation,PHYTOCHROMEINTERACTINGFACTOR3-LIKE1(PIL1)andXYLOGLUCANENDOTRANSGLYCOSYLASE7(XTR7),wasincreasedintheVQ29-OEplantsunderfar-redlightcondition(Figure2F).
Takentogether,theseresultsdemonstratethatVQ29isanegativeregulatorofseedlingde-etiolationunderfar-redandlowwhitelightconditions.
SubcellularlocalizationofVQ29TodeterminethesubcellularlocalizationofVQ29,wefirstfusedVQ29withgreenfluorescenceprotein(GFP)andtransientlyexpressedthisconstructinArabidopsisprotoplasts.
TheVQ29-GFPfusionproteinwasdetectedwithmultiplebandsusingtheGFPantibody,likelyduetopartialdegradationofthefusionproteininvitro(SupplementalFigureS2).
ConfocalmicroscopyrevealedthattheVQ29-GFPfusionwaslikelylocalizedtothenucleus,cytoplasmandplasmamembrane(Figure3A).
Furthermore,wegeneratedstabletransgenicplantsexpressingatranslationalfusionofVQ29withGFPunderthecontroloftheCaMV35Spromoter(Pro35S:VQ29-GFP).
GFPfluorescencewasdistributedmainlyinthenucleusofhypocotylcellsfromthesetransgenicplants(Figure3B).
TosubstantiatethelocalizationofVQ29,weisolatedproteinfractionsfromnucleus,cytoplasmandplasmamembraneofPro35S:Myc-VQ29(line#10)transgenicplants.
AsshowninFigure3C,Myc-VQ29fusionproteinwasonlydetectedincellfractionsisolatedfromthenucleus,butnotfromplasmamembraneorcytosol.
ItslocalizationinthecytoplasmandplasmamembraneintheprotoplastsinFigure3Amightbecausedbyover-expressionofVQ29-GFPintheprotoplasts.
ExpressionpatternofVQ29ToexamineVQ29expressionindifferenttissues,weanalyzeditstranscriptlevelsusingRT-PCR.
RelativelyhighlevelsofVQ29transcriptwereobservedinthestem,https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
12whereasexpressionwaslowintheroot,rosetteleaf,flower,andsilique(Figure4A).
TofurthervisualizetheexpressionpatternofVQ29,wegeneratedtransgenicplantsexpressingtheβ–glucuronidase(GUS)reportergeneunderthecontroloftheVQ29promotersequence(2.
0kbupstreamoftheATGtranslationalstartcodon).
HistochemicalstainingshowedthattheGUSreportergenewasexpressedintheradicle,hypocotyl,stem,leafvein,flower,andsiliquebase,indicatingthatVQ29mayfunctioninthesetissues(Figure4B).
WethenaskedwhetherVQ29isregulatedbylight.
phyA,phyB,andcry1photoreceptormutants,alongwiththeColwildtype,weregrowninfar-red,red,andbluelightconditions,respectively,for5d.
AsshowninFigure4C,quantitativeRT-PCRassaysshowedthatVQ29expressionwasrepressedbyvariouslighttreatments.
Furthermore,theVQ29transcriptlevelswereup-regulatedinthephyA-211andphyB-9mutantsunderfar-redandredlightconditionscomparedwiththewildtype,respectively.
NopronounceddifferenceinVQ29expressionwasfoundbetweenthecry1mutantandthewildtype.
Surprisingly,inthedark,VQ29expressionwasincreasedinphyBmutant,butwasnotaffectedbyphyAandcry1mutations,suggestingthatphyBplaysaroleinregulatingVQ29intheetiolatedseedlings(Figure4C).
TheseobservationssuggestthatlightinhibitsVQ29expressioninaphytochrome-dependentmanner,consistentwithitsroleinregulatingphotomorphogenesisunderfar-redandlowintensitylightconditions.
However,animmunoblottingassayusingtheMYCantibodyandtheVQ29-OEplantsshowedthattheproteinlevelofVQ29wasnotregulatedbylight(SupplementalFigureS5).
VQ29physicallyinteractswithPIF1PreviousstudiesreportedthatVQproteinscouldinteractwithvariousWRKYTFs(Huetal.
,2013,Laietal.
,2011,Wangetal.
,2010).
ThefindingsthatVQ29possessestranscriptionalactivityandisinvolvedinregulatingphotomorphogenesispromptedustohypothesizethatVQ29activitymightdependonitsinteractionwithotherTF(s).
SincePIFproteins,includingPIF1,3,4and5,arebHLH-typeTFsthatplayacriticalroleinrepressingphotomorphogenesis(Leivaretal.
,2008,Shinetal.
,https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
132009),wetestedtheirpossibleinteractionwithVQ29inayeasttwo-hybridsystem.
VQ29wasfusedwiththeLexADNA-bindingdomain(LexA-VQ29)andvariousPIFproteinswereligatedwiththeactivationdomainofB42(AD-PIFs).
Co-expressionofLexA-VQ29withAD-PIF1orAD-PIF3causedstrongactivationoftheLacZreporter(Figure5A,B),indicatingthatVQ29interactswithPIF1andPIF3inyeastcells.
However,onlymildinteractionwasdetectedbetweenVQ29andPIF5.
Asnegativecontrols,AD-SIG1(plastid-locatedSIGMAFACTOR1)orADalonefailedtointeractwithLexA-VQ29(Figure5A,B).
Inthisstudy,therelationshipbetweenVQ29andPIF1wasfurtheranalyzed.
TosubstantiatetheinteractionbetweenVQ29andPIF1inplantcells,weperformedabimolecularfluorescencecomplementation(BiFC)assayinwhichwetransientlyco-expressedtheN-terminusofyellowfluorescenceproteinfusedtoVQ29(YFPN-VQ29)andPIF1fusedtotheC-terminusofYFP(PIF1-YFPC)inArabidopsisprotoplasts(Walteretal.
,2004).
Co-expressionofYFPN-VQ29andPIF1-YFPCreconstitutedafunctionalYFPinthenucleus,whereasco-expressionwitheithercontrolvectorfailedtogenerateYFPfluorescence(Figure5C).
TheinvivointeractionbetweenVQ29andPIF1wasfurtherconfirmedbyco-immunoprecipitationassay.
ThereforewegenerateddoubletransgenicplantsPro35S:VQ29-GFP/Pro35S:TAP-PIF1bycrossingtheirsingletransgenicplant.
TAP-PIF1(usingMYCantibody)wasabletopulldownVQ29-GFP(detectedbyGFPantibody)inthedoubletransgenicseedlings(Figure5D).
Inaddition,twopointmutationsintheVQmotifofVQ29(V70D,Q71L)didnotaffectitsinteractionwithPIF1(Figure5C).
ThesedatatogetherindicatethatVQ29indeedinteractswithPIF1andthattheVQmotifislikelynotrequiredformediatingtheinteraction.
VQ29andPIF1co-regulateseedlingde-etiolationThephysicalinteractionbetweenVQ29andPIF1ledustoinvestigatewhetherandhowtheseproteinsfunctiontogetherinthelightsignalingpathwayinArabidopsis.
Tothisend,wegeneratedthevq29pif1doublemutantbycrossingvq29-1andpif1-2mutants,andVQ29-OEPIF1-OEdoubletransgenicplantsbycrossinghttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
14Pro35S:Myc-VQ29(line#10)andPro35S:TAP-PIF1.
Weexaminedthehypocotylelongationphenotypesofthedoublehomozygouslines,theirsinglemutantortransgenicparentplants,andthewildtype.
Thehypocotyllengthofthevq29pif1doublemutantwasshorterthanthatofthesinglemutantsandColwild-typeseedlingsunderbothfar-redandlowlightconditions(Figure6A,B,SupplementalFigureS3).
Furthermore,thevq29pif1doublemutantwaspronouncedlyshorterthanthesinglemutantsunderfar-red/darkcycles,orunderfar-redlightconditionswithoutsucrosesupplementinthemedia(Figure6C,SupplementalFigureS4B).
Bycontrast,theVQ29-OEPIF1-OEdoubletransgenicplantsdisplayedmuchlongerhypocotylsthantheirparentsingleoverexpressionlinesandthewild-typeplantsunderlowlightconditions(Figure6D).
ThesedatatogetherindicatethatVQ29andPIF1additivelyrepressphotomorphogenesis.
SincePIF1isalsoinvolvedinregulatingphytochrome-mediatedseedgerminationandseedlinggrowthduringde-etiolation(Ohetal.
,2004;Leivaretal.
,2008;Huqetal.
,2004,),wenexttestedwhethermutationoroverexpressionofVQ29havetheseresponses.
Aspreviouslyreported,pif1mutantdisplayedhighgerminationrateafter5minoffar-redlighttreatment(SupplementalFigureS6A).
Seedsofvq29mutantandVQ29-OEdidnotshowanygerminationdifferencecomparedtowildtype,andthegerminationefficiencyofvq29pif1doublemutantwassimilaraspif1(SupplementalFigureS6A).
Inaddition,mutationoroverexpressionofVQ29didnotshowseedlinggreeningphenotype,andvq29-1didnotaffectthedefectsofpif1mutantduringde-etiolation(SupplementalFigureS6B).
TheseobservationsindicatethatVQ29isnotinvolvedinseedgerminationandseedlinggreeningresponseswithPIF1,andthatitsresponsivenessinhypocotylelongationappearstobespecific.
VQ29bindstothepromotersofPIL1andXTR7andregulatestheirexpressionwithPIF1PIF1alsoregulatestheexpressionoflight-responsiveandcellelongation-relatedgenes,suchasPIL1andXTR7(Leivaretal.
,2012).
QuantitativeRT-PCRanalysisrevealedthattheexpressionofPIL1andXTR7wasgreatlydown-regulatedinhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
15darknessinthevq29pif1doublemutantcomparedwithitsparentmutantsandthewildtype(Figure7A).
Inaddition,thetranscriptlevelsofanothertwogenesinvolvedincellelongation,EXTENSIN1(EXT1)andEXT3,weredecreasedinvq29andpif1mutants,andfurtherreducedinthevq29pif1doublemutant(SupplementalFigureS7).
WealsotestedtheexpressionofEXPANSIN8(EXP8)andEXP10,twogenesthatwererepressedbyPIF1(Ohetal.
,2009).
SurprisinglytheEXP10transcriptlevelwasincreasedinvq29pif1doublemutantcomparedthatinpif1(SupplementalFigureS7).
Furthermore,vectorsharboringtheLUCreportergenedriveneitherbythePIL1orXTR7promoterwereconstructedandco-transformedwithVQ29and/orPIF1intoArabidopsisprotoplasts.
AsshowninFigure7B,VQ29itselfdidnotsignificantlypromoteLUCexpressionunderthecontrolofeitherthePIL1orXTR7promoter,whereasPIF1alonestronglyactivatedLUCexpression.
Mostremarkably,co-expressionofPIF1andVQ29increasedtheexpressionlevelsoftheseLUCreportergenes.
Interestingly,amutatedversionofVQ29(V70D)furtheractivatedthelevelsofLUCexpressionwhenco-transformedwithPIF1(Figure7B),suggestingthattheVQdomainofVQ29indeedpossessestranscriptionrepressionfunction.
Consistentwiththeseobservations,theexpressionofPIL1andXTR7wasincreasedinPIF1-OEtransgenicplants,andXTR7'slevelwasfurtheractivatedinVQ29-OEPIF1-OEdoubletransgenicplants(Figure7C).
Therefore,PIF1andVQ29coordinatelyregulatedownstreamgeneexpression,andVQ29likelystimulatestheactivationactivityofPIF1.
ToassesswhethertheinductionofPIL1andXTR7isdirectlyaffectedbyPIF1andVQ29,wecarriedoutchromatinimmunoprecipitationfollowedbyquantitativePCR(ChIP-qPCR)assays.
WhenprecipitatedwithGFPantibodies,fragmentsofthePIL1andXTR7promoterscontainingtheG-box(P),butnottheircodingregions(C),weregreatlyenrichedinextractsfromPro35S:VQ29-GFPtransgenicseedlings,butnotfromColwild-typeseedlings(Figure7D,E).
Inaddition,MYCantibodywasabletopulldownthepromoterfragmentsofXTR7,butnotofPIL1,inextractsfromPro35S:TAP-PIF1transgenicplants(Figure7F).
ThesedataindicatethatVQ29isassociatedwiththepromoterregionsofPIL1andXTR7inplantcells.
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
16PIF1israpidlydegradedafterlighttransition(Shenetal.
,2005;2008).
TotestwhetherVQ29couldaffectthestabilityofPIF1,weperformedanimmunoblottingassayusingtheMYCantibody,andPIF1-OEandPIF1-OEVQ29-OEtransgenicplants.
OurresultshowedthatoverexpressingVQ29didnotaffectthestabilityanddegradationofPIF1(SupplementalFigureS8).
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
17DISCUSSIONVQ29definesanovelrepressorofseedlingde-etiolationInthisstudy,wepresentmultiplelinesofevidencethatVQ29isanoveltranscriptionregulatorthatrepressesseedlingde-etiolationinArabidopsis.
First,hypocotylelongationinplantsoverexpressingVQ29exhibitsreducedsensitivityspecificallytofar-redandlowlightconditions(Figure2).
Itshouldbenotedthatalthoughnoeffectofvq29-1mutantinfar-redlightwasobservedinFigures2Eand6B,astatisticallysignificantshorthypocotylphenotypewasobservedinthismutantinlowfar-redlightintensities(SupplementalFigureS3)orinfar-redlightwithoutsucrose,inmultipleindependentexperiments(SupplementalFigureS4).
Theseoppositephenotypesoftheloss-of-functionmutantandoverexpressionplantsofVQ29suggestthatitscodingproteinisinvolvedinthephytochromesignalingpathway.
Inagreementwiththis,VQ29expressionisdown-regulatedbylightinaphytochrome-dependentmanner(Figure4C).
Second,yeasttwo-hybrid,BiFCandCo-IPassaysshowedthatVQ29isabletophysicallyinteractwithPIF1,akeytranscriptionfactorinthelightsignalingpathway,bothinyeastandinplantcells(Figure5).
Third,thefactsthatthevq29pif1doublemutantorVQ29-OEPIF1-OEdoubletransgenicplantsexhibitedpronouncedphenotypescomparedwiththeircorrespondingsingleparentplantssuggestthatVQ29andPIF1haveadditiveeffect(Figure6,SupplementalFigureS3,S4).
Fourth,VQ29andPIF1functiontogethertoactivatetheexpressionoffourgenesinvolvedinpromotingcellelongation.
Furthermore,thisregulationmightbedirect,asVQ29isassociatedwiththepromotersofPIL1andXTR7invivo(Figure7E).
Thelessstrongphenotypeinthevq29mutantcouldbealternativelycausedbythefunctionalredundancyofVQ29withotherrelatedVQgenes.
FuturestudiesusingdoubleorhighordermutantswithotherVQsshouldfurthertestthispossibility.
PIF1isstabilizedindarknessanddegradedbylight(Shenetal.
,2005,2008).
ItdirectlybindstoDNAviatheG-boxcis-elementinthepromoterofdownstreamgenesandregulatestheirexpression(LeivarandQuail,2010).
Consistently,aG-boxmotifisfoundinthepromoterregionsofbothPIL1andXTR7.
Therefore,weproposehttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
18thatVQ29transcriptaccumulatesindarknessanditscodingproteinactsasatranscriptionco-regulatortopromotetheactivityofPIF1throughphysicalinteraction,leadingtotheactivationofcellelongation-relatedgenes(e.
g.
,PIL1andXTR7),therebyinhibitingseedlingde-etiolation(Figure8).
Infar-redlightconditions,thelevelsofPIF1proteinandVQ29transcriptarereduced,resultingintheinhibitionofcellelongation-relatedgeneexpression,andconsequentlythepromotionofphotomorphogenesis.
Alternatively,VQ29mightfunctionasatranscriptionfactorthatdirectlybindstothepromoterofdownstreamgenesindependentofPIF1.
Furtherstudyisdeservedtoexaminethispossibility.
PIL1(encodesaPIF-regulatedtranscriptionfactor)andXTR7(encodesaxyloglucanendotransglycosylase-relatedprotein)areearlyshademarkergenes(Hornitscheketal.
,2009)thataredirectlyregulatedbyPIF1andVQ29.
VQ29isrequiredforregulatingphotomorphogenesisspecificallyinlowwhitelightandfar-redlight,whichmimictheshadeconditions.
Hence,theinvolvementofVQ29mightfinetunetheresponsivenesstoshade-likeenvironmentbycoordinatingwithPIF1.
Consistently,PIF1alsoplaysarole,althoughmoderate,inshadeavoidanceresponse(Leivaretal.
,2012).
OurChIPexperimentshowedthatVQ29isassociatedwiththepromoterregionofXTR7.
However,nodifferenceintheexpressionofXTR7invq29-1mutantandwildtypewasfound,suggestingthatotherfactor(s)isrequiredforregulatingdownstreamgeneexpressionwithVQ29.
Inagreementwiththisnotion,XTR7mRNAlevelwasgreatlyreducedinthevq29pif1doublemutant.
Inaddition,itisalsopossiblethatotherVQmember(s)playsaroleinregulatingdownstreamgeneexpressionwithVQ29.
AccumulatingstudiessuggestthatmembersoftheVQfamilyplaydiversefunctions,suchasregulatingtheplantdefenseresponse(Narusakaetal.
,2008,Xieetal.
,2010,Laietal.
,2011,Andereassonetal.
,2005),abioticstressresistance(Perrucetal.
,2004,Huetal.
,2013),andseeddevelopment(Wangetal.
,2010).
OurstudyprovidesgeneticandmolecularevidencethatVQ29hasaprominentroleinplantgrowthanddevelopmentinresponsetothelightenvironment,thusextendingthefunctionalityofthisplant-specificproteinfamily.
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
19VQproteinsareinvolvedintranscriptionalregulationVQsaresmallproteinswithashort,uniqueVQmotif.
MolecularstudiesindicatethatVQproteinslikelyinteractwithWRKYtranscriptionfactorsandotherproteins,suchaskinasesandplastidsigmafactors(Laietal.
,2011,Wangetal.
,2010,Chengetal.
,2012,Huetal.
,2013,Andereassonetal.
,2005).
WRKYandPIFareplant-specifictranscriptionfactorsthatplaycrucialbiologicalroles(Rushtonetal.
,2010,LeivarandQuail,2010).
TheinteractionofVQtranscriptionregulatorswithothertranscriptionfactorsappearstofine-tunethetemporalandspatialexpressionpatternsofspecifictargetsduringplantdevelopment,oruponexposuretoparticularbioticandabioticstresses.
OurdatashowedthatmutationsintheVQdomainofVQ29donotaffecttheinteractionwithPIF1(Figure5C).
However,arecentstudyrevealedthattheVQmotifofSIB1isimportantforitsinteractionwithWRKY33(Laietal.
,2011).
Therefore,thereisvariationinwhichdomainsofVQproteinsinteractwithotherproteins.
VQ29possessestranscriptionalrepressionactivityinatransientexpressionsystem.
OurdetailanalysisoftheconservedVandQresiduesintheVQmotifindicatesthatpointmutationsofVQ29alteritstranscriptionalactivity(Figure1D).
Moreover,mutationinVQ29(V70D)furtheractivatesProPIL1:LUCandProXTR7:LUCreporterexpressionmediatedbyPIF1(Figure7B).
TheseobservationsdemonstratethattheVQdomainofVQ29indeedhasrepressivefunctionongeneexpression.
However,VQ29helpsPIF1topromotePIF1-inducedgeneexpression,andtorepresstheexpressionofPIF1-inhibitedgenesduringlight-mediatedearlyseedlinggrowth.
Hence,theactivityofVQ29anditseffectongeneexpressionlikelydependonspecificinteractingfactorandthetargetgenesinagivensignalingprocess.
Similarly,SIB1simulatestheDNA-bindingandtranscriptionalactivityofWRKY33intheplantdefenseresponse(Laietal.
,2011).
Incontrast,recruitmentofVQ9leadstotheinactivationofWRKY8,andtheseproteinsactantagonisticallytomediatethesaltstressresponse(Huetal.
,2013).
OurtransientexpressionassayinprotoplastssuggeststhatthemajorityofVQproteinsexhibiteithertranscriptionalactivationorrepressionactivities(Figure1).
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
20ThefactthatVQ29localizestothenucleusfurthersupportsitsroleinregulatingtranscription.
Furthermore,almostallmembersofthisfamilyarethoughttolocalizetothenucleus,basedonsubcellularproteomicdatabasepredictions(Chengetal.
,2012).
Theidentificationofmoreinteractingproteins,particularlyoftranscriptionfactorsinthenucleus,shouldfurtherelucidatethefunctionsandmechanismsofVQproteinsinplants.
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
21MATERIALSANDMETHODSPlantmaterialsandgrowthconditionsThevq29-1(Salk_061586andSalk_061438),pif1-2,phyA-211,phyB-9andcry1-304mutantsandPro35S:TAP-PIF1transgenicplantswerederivedfromtheArabidopsisthalianaColumbia(Col)ecotype(Chenetal.
,2013,Reedetal.
,1994,1993,Mockleretal.
,1999,Moonetal.
,2008).
vq29-1wasconfirmedbyPCRgenotypingandtheT-DNAinsertionsitewasverifiedbysequencing.
Doublemutantortransgenicplantsweregeneratedbygeneticcrossingandhomozygouslineswereusedintheseexperiments.
SeedlingsweregrownonMurashigeandSkoogmediumcontaining1%sucrose,orwithoutsucroseasindicatedinthetextandSupplementalFigureS4.
Far-redlight(12molm-2s-1),redlight(20molm-2s-1),andbluelight(14molm-2s-1)weresuppliedbylight-emittingdiodelightsources,andlowwhitelight(10molm-2s-1)wassuppliedbycoolwhitefluorescentlamps.
Forfluencerateanalysis,thelightintensitieswereindicatedinthefigures.
DeterminationofHypocotylLength,SeedlingGreeningandSeedGerminationRateForhypocotyllengthmeasurement,seedlingsofdifferentgenotypesweregrownsidebysideonthesameplate,andatleastthreeindependentplateswereusedinallexperiments.
SeedlingswerethenphotographedandtheirhypocotyllengthwasmeasuredbyusingImageJsoftware(http://rsb.
info.
nih.
gov/ij).
Forseedlinggreeningrateanalysis,dark-grownseedlingsweretransferredtocontinuouswhitelightfor2d.
Greeningratewasdeterminedbycountingthepercentageofdark-greencotyledonsfrom50to80seedlingsofeachgenotype.
Forseedgerminationassay,seedsofdifferentgenotypeswereharvestedonthesamedayfromplantsgrowninidenticalconditions.
Seedgerminationwasobservedunderamicroscopeanddeterminedbasedontheappearanceofradicleprotrusion.
Geneexpressionanalysishttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
22PlanttotalRNAwasextractedusinganRNAprepPurePlantKit(Tiangen),andfirststrandcDNAwassynthesizedusingReverseTranscriptase(Invitrogen).
Real-timePCRwascarriedoutusingtheSYBRPremixExTaqKit(Takara)followingthemanufacturer'sinstructions.
Theexpressionlevelswerenormalizedtotheexpressionofaubiquitin(UBQ1)gene.
PrimersarelistedinSupplementalTableS1.
PlasmidconstructionToobtainopenreadingframes(ORFs)ofVQ2,PIF4,andPIF5,firststrandcDNAwasreversetranscribedfromtotalRNAextractedfromColwild-typeseedlingsusingoligo(dT)18primerandhighfidelityPfuDNApolymerase(Invitrogen).
TheproductswereclonedintothepEASYvector(TransGen),resultinginpEASY-VQ2,pEASY-PIF4,andpEASY-PIF5,respectively.
DuetotheabsenceofintronsinallVQgenes(exceptVQ2),theORFswereamplifiedfromgenomicDNAusingPfuandclonedintothepEASYvector,resultinginpEASY-VQs,respectively.
MutationsinVQ29(V70A,V70D,Q71L,V70DQ71L)weregeneratedusingaMutantBESTsite-directedmutagenesiskit(Takara)accordingtothemanufacture'sinstructions.
Appropriaterestrictionenzymesitesweredesignedattheendofeachprimer(SupplementalTableS1).
AllamplifiedORFswerevalidatedbysequencing.
Togenerateconstructsfortheprotoplasttransientexpressionassay,theVQ1fragmentwasreleasedfrompEASY-VQ1digestedwithBamHIandXhoI,andinsertedintotheBglII/SalIsiteofpSAT-GAL4DB(Jingetal.
,2013),toyieldpGAL4DB-VQ1.
TheORFsofVQ4,VQ5,VQ6,VQ7,andVQ18werereleasedfromthecorrespondingMfeI-XhoI-digestedpEASY-VQvectors,respectively,andclonedintotheEcoRI/SalIsitesofpSAT-GAL4DB,givingrisetopGAL4DB-VQ4/5/6/7/18.
TheORFsofVQ24andVQ30werereleasedfrompEASY-VQ24/30digestedwithEcoRIandSalI,andinsertedintotheEcoRI/SalIsitesofpSAT-GAL4DB,generatingpGAL4DB-VQ24andpGAL4DB-VQ30,respectively.
ToobtainpGAL4DB-VQ34,thepEASY-VQ34plasmidwascutwithMfeIandSalI,andtheVQ34ORFwasclonedintotheEcoRI/SalIsitesofpSAT-GAL4DB.
TheORFsofVQ2/3/8/9/10/11/12/13/14/15/16/17/19/20/21/22/23/25/26/27/28/29/31/32/33werehttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
23releasedfromthecorrespondingEcoRI-XhoI-digestedpEASY-VQvectors,respectively,andinsertedintotheEcoRI/SalIsitesofpSAT-GAL4DB,resultinginthecorrespondingpGAL4DB-VQsconstructs.
Toconstructvectorsfortheyeasttwo-hybridassay,thepEASY-VQ29plasmidwascutwithEcoRIandXhoI,andtheVQ29fragmentwasligatedintoEcoRI/XhoI-digestedEG202vector(Clontech),togiverisetopLexA-VQ29.
ThepAD-PIF1andpAD-PIF3plasmidswereconstructedinapreviousstudy(Chenetal.
,2013).
ThepEASY-PIF4andpEASY-PIF5plasmidsweredigestedwithEcoRIandSalI,andthePIF4orPIF5fragmentswereinsertedintotheEcoRI/XhoIsitesoftheJG4-5vector(Clontech),resultinginpAD-PIF4orpAD-PIF5,respectively.
ToprepareconstructsfortheBiFCassay,theVQ29fragmentreleasedfromEcoRI/XhoI–digestedpEASY-VQ29wasinsertedintothepUC-SPYNEvector(Walteretal.
,2004)digestedwithEcoRIandXhoI,togeneratepYFPN-VQ29.
pYFPC-PIF1wasdescribedinapreviousstudy(Chenetal.
,2013).
TostudythesubcellularlocalizationofVQ29,thefragmentreleasedfrompEASY-VQ29usingEcoRIandXhoIwasligatedintotheEcoRI/SalIsitesofthemodifiedpdGNvector(Leeetal.
,2005),togeneratepGFP-VQ29.
ToconstructProVQ29:GUS,afragmentspanningtheregion2-kbupstreamoftheATGstartsiteofVQ29wasamplifiedbyPCRandclonedintopEASY,resultinginpEASY-VQ29p.
ThepEASY-VQ29pplasmidwasthendigestedwithSalIandBamHItoreleasetheVQ29promoterfragment,whichwasthenligatedintothepBI101vectordigestedwiththesameenzymes,togenerateProVQ29:GUS.
ToconstructVQ29overexpressionvectors,theVQ29codingsequencewasreleasedfrompEASY-VQ29usingEcoRIandXhoI,andthenligatedintotheEcoRI-SalIsitesofmodifiedpRI101(Takara),inwhichthreecopiesofMYCtagwereinserted,resultinginPro35S:Myc-VQ29.
TheVQ29ORFwasamplifiedfrompEASY-VQ29byintroducinganNcoIsiteattheN-terminalandaSpeIsiteattheC-terminal,andclonedintopEASYtoproducepEASY-VQ29G.
ThepEASY-VQ29GplasmidwasthendigestedwithNcoIandSpeIandtheVQ29fragmentwasinsertedintopCAMBIA1302(www.
cambia.
org/daisy/cambia/585)cuthttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
24withthesameenzymes,togeneratePro35S:VQ29-GFP.
TogenerateconstructsforthetransientexpressionofPIL1andXTR7,thepromotersequencesofbothgeneswereamplifiedfromColgenomicDNAandligatedintothepEASYvector,resultinginpEASY-PIL1pandpEASY-XTR7p,respectively.
TheseplasmidswerecutwithHindIIIandBamHItoreleasethePIL1andXTR7fragments,whichwereinsertedintotheHindIII/BamHIsitesofthepUC-35sLUCvector(Chenetal.
,2013)toproduceProPIL1:LUCandProXTR7:LUC,respectively.
ThebinaryconstructswereelectroporatedintoAgrobacteriumtumefaciensstrainGV3101andthenintroducedintoColwildtypeplants.
TransgenicplantswereselectedonMSplatesinthepresenceof50mg/Lkanamycinorhygromycin,andhomozygouslineswereusedinthisstudy.
LuciferasetransientexpressionassayTheLUCtransientexpressionassaywascarriedoutaspreviouslydescribed(Jingetal.
,2013).
LUCreporteractivitywasdetectedwithaluminescencekitusingLuciferaseAssaySystem(Promega)andtherelativeactivitywasexpressedastheLUC/GUSratios.
GUShistochemicalassaySeedlingsoftheProVQ29:GUStransgeniclinewereharvestedandincubatedovernightin0.
1Msodiumphosphatebuffercontaining50mMK3Fe(CN)6,50mMK4Fe(CN)6,and1mM5-bromo-4-chloro-3-indolyl-β-D-glucuronideat37°C.
GUSexpressionwasexaminedunderadissectingmicroscopeandimageswerecapturedbyadigitalcamera(Olympus).
GFPproteinlocalizationassayForthetransientassay,VQ29-GFPwastransformedintoArabidopsisprotoplastsandtheprotoplastswereincubatedunderdarknessfor16hbeforeobservation.
ForGFPlocalizationinstabletransgenicplants,ahomozygouslineofPro35S:VQ29-GFPwasused.
TheprotoplastsandtransgenicseedlingsweremountedonaslideandGFPhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
25fluorescencewasvisualizedwithaLeicaTCSSP5confocalmicroscope.
Yeasttwo-hybridassayYeasttwo-hybridanalysiswasperformedaspreviouslydescribed(Tangetal.
,2012).
TransformantsweregrownonSD/-Trp-Ura-HisdropoutplatescontainingX-gal(5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside)forcolordevelopment.
Relativebeta-galactocidaseactivitywasquantifiedaccordingtothemethoddescribedbyYeastProtocolsHandbook(Clontech).
Co-IPandImmunoblottingSeedlingswerehomogenizedinextractionbuffercontaining50mMTris-HCl,pH7.
5,150mMNaCl,10mMMgCl2,0.
1%Tween20,1mMphenylmethylsulfonylfluoride,and1*completeproteaseinhibitorcocktail(Roche).
Theextractswerecentrifugedat14,000*gtwiceat4°Cfor10mineach,andproteinconcentrationwasdeterminedusingtheBradfordassay(Bio-Rad).
Proteinfractionationsfromnucleus,plasmamembraneorcytoplasmwereisolatedaccordingtothemethodsasdescribed(Larssonetal.
,1994,Liuetal.
,2001).
Co-IPassaywascarriedoutaspreviouslydescribed(Tangetal.
,2012).
Proteinswereseparatedon10%SDS-PAGEgelsandtransferredontopolyvinylidenefluoridemembranes.
Theywerethenblottedagainstanti-MYC(Abcam),anti-GFP(Sigma),anti-H+-ATPase(Agrisera),anti-cFBPase(Agrisera),oranti-tubulin(Jingetal.
,2013)antibodies.
Theproteinbandswerevisualizedusingthestandardenhancedchemiluminescencemethod.
BiFCPlasmidscontainingN-andC-terminalYFPfusionswereco-transformedintoArabidopsisprotoplastsaspreviouslydescribed(Walteretal.
,2004).
Theprotoplastswereincubatedunderweaklightfor12-16hbeforeobservation.
YFPfluorescencewascapturedwithaconfocalmicroscope(Leica).
ChIPhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
26TheColwildtypeandPro35S:TAP-PIF1andPro35S:VQ29-GFPtransgenicplantswereusedintheChIPassay,whichwascarriedoutaspreviouslydescribed(Chenetal.
,2013).
Thechromatincomplexeswereincubatedwithanti-MYCoranti-GFPpolyclonalantibodies.
TheprecipitatedDNAfragmentswererecoveredandquantifiedbyquantitativePCRwiththeprimersshowninSupplementalTableS1.
AccessionNumbersSequencedatafromthisarticlecanbefoundintheArabidopsisGenomeInitiativeorGenBank/EMBLdatabasesunderthefollowingaccessionnumbers:VQ29(At4g37710),PIF1(At2g20180),PIF3(At1g09530),PIF4(At2g43010),PIF5(At3g59060),PIL1(At2g46970),XTR7(At4g14130),EXT1(At1g76930),EXT3(At1g21310),EXP8(At2g40610),EXP10(At1g26770),SIG1(At1g08540),ACTIN(At3g18780)andUBQ1(At3g52590),andtheaccessionnumbersofallotherVQgenesarelistedinSupplementalTableS1.
SupplementalDataThefollowingmaterialsareavailableintheonlineversionofthisarticle.
SupplementalFigureS1.
ComparisonofVQgenesinArabidopsis,rice,andmoss.
SupplementalFigureS2.
VQ29proteinlevelinwildtypeandvariouspointmutants.
SupplementalFigureS3.
Fluence-rateresponseunderdifferentlightconditions.
SupplementalFigureS4.
HypocotyllengthofVQ29mutantandoverexpressionplantsgrowninthemediawithoutsucrose.
SupplementalFigureS5.
VQ29proteinlevelduringdark-to-lighttransition.
SupplementalFigureS6.
Phenotypeinseedlinggreeningandseedgermination.
SupplementalFigureS7.
Downstreamgeneexpressioninvq29and/orpif1mutants.
SupplementalFigureS8.
TheeffectofVQ29onPIF1proteinstability.
SupplementalTableS1.
Listofprimersusedinthisstudy.
ACKNOWLEDGMENTShttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
27WethanktheArabidopsisBiologicalResourceCenterforprovidingthevq29mutant.
WearegratefultoDr.
EnamulHuq(UniversityofTexasatAustin)forprovidingthemutantandoverexpressiontransgenicseedsofPIF1,andDr.
TaiWang(InstituteofBotany,ChineseAcademyofSciences)forgiftingplasmamembrane-andcytosol-localizedantibodies.
LITERATURECITEDAndreassonE,JenkinsT,BrodersenP,ThorgrimsenS,PetersenNHT,ZhuS,QiuJL,MicheelsenP,RocherA,PetersenM,NewmanMA,NielsenHB,HirtH,SomssichI,MattssonO,MundyJ(2005)TheMAPkinasesubstrateMKS1isaregulatorofplantdefenseresponses.
EMBOJ24:2579-2589ArsovskiAA,GalstyanA,GusemanJM,NemhauserJL(2012)Photomorphogenesis.
ArabidopsisBook,10:e0147.
doi:10.
1199/tab.
0147BallesterosML,BolleC,LoisLM,MooreJM,Vielle-CalzadaJP,GrossniklausU,ChuaNH(2001)LAF1,aMYBtranscriptionactivatorforphytochromeAsignaling.
GenesDev15:2613–2625ChangCJ,MaloofJN,WuSH(2011)COP1-mediateddegradationofBBX22/LZF1optimizesseedlingdevelopmentinArabidopsis.
PlantPhysiol156:228-239ChenD,XuG,TangW,JingY,JiQ,FeiZ,LinR(2013)AntagonisticbHLH/bZIPtranscriptionfactorsformtranscriptionalmodulesthatintegratelightandreactiveoxygenspeciessignalinginArabidopsis.
PlantCell25:1657-1673ChenM,ChoryJ,FankhauserC(2004)Lightsignaltransductioninhigherplants.
AnnuRevGenet38:87-117ChengY,ZhouY,YangY,ChiYJ,ZhouJ,ChenJY,WangF,FanB,ShiK,ZhouYH,YuJQ,ChenZ(2012)StructuralandfunctionalanalysisofVQmotif-containingproteinsinArabidopsisasinteractingproteinsofWRKYtranscriptionfactors.
PlantPhysiol159:810-825ChoiG,YiH,LeeJ,KwonYK,SohMS,ShinB,LukaZ,HahnTR,SongPS(1999)Phytochromesignalingismediatedthroughnucleosidediphosphatekinasehttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
282.
Nature401:610–613DattaS,HettiarachchiC,JohanssonH,HolmM(2007)SALTTOLERANCEHOMOLOG2,aB-boxproteininArabidopsisthatactivatestranscriptionandpositivelyregulateslight-mediateddevelopment.
PlantCell19:3242-3255DieterleM,ZhouYC,SchaferE,FunkM,KretschT(2001)EID1,anF-boxproteininvolvedinphytochromeA-specificlightsignaling.
GenesDev15:939-944HarmonFG,KaySA(2003)TheFboxproteinAFRisapositveregulatorofphytochromeA-mediatedsignaling.
CurrBiol13:2091-2096HornitschekP,LorrainS,ZoeteV,MichielinO,FankhauserC(2009)Inhibitionoftheshadeavoidanceresponsebyformationofnon-DNAbindingbHLHheterodimers.
EMBOJ28:3893-3902HsiehWP,HsiehHL,WuSH(2012)ArabidopsisbZIP16transcriptionfactorintegrateslightandhormonesignalingpathwaystoregulateearlyseedlingdevelopment.
PlantCell24:3997-4011HuY,ChenL,WangH,ZhangL,WangF,YuD(2013)ArabidopsistranscriptionfactorWRKY8functionsantagonisticallywithitsinteractingpartnerVQ9tomodulatesalinitystresstolerance.
PlantJdoi/10.
1111/tpj.
12159HudsonM,RingliC,BoylanMT,QuailPH(1999)TheFAR1locusencodesanovelnuclearproteinspecifictophytochromeAsignaling.
GenesDev13:2017–2027HuqE,Al-SadyB,HudsonM,KimCH,ApelM,QuailPH(2004)PHYTOCHROME-INTERACTINGFACTOR1isacriticalbHLHregulatorofchlorophyllbiosynthesis.
Science305:1937-1941IwataY,FedoroffNV,KoizumiN(2008)ArabidopsisbZIP60isaproteolysis-activatedtranscriptionfactorinvolvedintheendoplasmicreticulumstressresponse.
PlantCell20:3107–3121.
JiaoY,LauOS,DengXW(2007)Light-regulatedtranscriptionalnetworkinhigherplants.
NatRevGene8:217-230JingY,ZhangD,WangX,TangW,WangW,HuaiJ,XuG,ChenD,LiY,LinRhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
29(2013)Arabidopsischromatin-remodelingfactorPICKLEinteractswithtranscriptionfactorHY5toregulatehypocotylcellelongation.
PlantCell25:242-256LaiZ,LiY,WangF,ChengY,FanB,YuJQ,ChenZ(2011)ArabidopsissigmafactorbindingproteinsareactivatorsoftheWRKY33transcriptionfactorinplantdefense.
PlantCell23:3824-3841LarssonC,SommarinM,WidellS(1994)Isolationofhighlypurifiedplantplasmamembranesandseparationofinside-outandright-side-outvesicles.
MethodsEnzymol228:451-469LauOS,DengXW(2010)Planthormonesignalinglightensup:integratorsoflightandhormones.
CurrOpinPlantBiol13:571-577LauOS,DengXW(2012)ThephotomorphogenicrepressorsCOP1andDET1:20yearslater.
TrendsPlantSci17:584-93LeeJ,TaokaK,YooB,Ben-NissanG,KimD,LucasWJ(2005)Plasmodesmal-associatedproteinkinaseintobaccoandArabidopsisrecognizesasubsetofnon-cell-autonomousproteins.
PlantCell17:2817-2831LeivarP,MonteE,OkaY,LiuT,CarleC,CastillonA,HuqE,QuailPH(2008)Multiplephytochrome-interactingbHLHtranscriptionfactorsrepressprematureseedlingphotomorphogenesisindarkness.
CurrBiol18:1815-1823LeivarP,QuailPH(2010)PIFs:pivotalcomponentsinacellularsignalinghub.
TrendsPlantSci16:19-28LeivarP,TeppermanJM,CohnMM,MontteE,Al-SadyB,EricksonE,QuailPH(2012)DynamicantagonismbetweenphytochromesandPIFfamilybasichelix-loop-helixfactorsinducesselectivereciprocalresponsestolightandshadeinarapidresponsivetranscriptionalnetworkinArabidopsis.
PlantCell24:1398-1419LiuX,CovingtonMF,FankhauserC,ChoryJ,WagnerDR(2001)ELF3encodesacircadianclock–regulatednuclearproteinthatfunctionsinanArabidopsisPHYBsignaltransductionpathway.
PlantCell13:1293-1304MocklerTC,GuoH,YangH,DuongH,Lin,C(1999)Antagonisticactionsofhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
30ArabidopsiscryptochromesandphytochromeBintheregulationoffloralinduction.
Development126:2073–2082MllerSG,KimYS,KunkelT,ChuaNH(2003)PP7isapositiveregulatorofbluelightsignalinginArabidopsis.
PlantCell15:1111–1119MoonJ,ZhuL,ShenH,HuqE(2008)PIF1directlyandindirectlyregulateschlorophyllbiosynthesistooptimizethegreeningprocessinArabidopsis.
ProcNatlAcadSciUSA105:9433-9438MorikawaK,ShiinaT,MurakamiS,ToyoshimaY(2002)Novelnuclear-encodedproteinsinteractingwithaplastidfactor,Sig1,inArabidopsisthaliana.
FEBSLetters514:300-304NarusakaM,KawaiK,izawaN,SekiM,ShinozakiK,SeoS,KobayashiM,ShiraishiT,NarusakaY(2008)GenecodingforSigA-bindingproteinfromArabidopsisappearstobetranscriptionallyup-regulatedbysalicylicacidandNPR1-dependentmechanisms.
JGenPlantPathol74:345-354OhE,KimJ,ParkE,KimJ,KangC,ChoiG(2004)PIL5,aphytochrome-interactingbasichelix-loop-helixprotein,isakeynegativeregulatorofseedgerminationinArabidopsisthaliana.
PlantCell16:3045-3058OhE,KangH,YamaguchiS,ParkJ,LeeD,KamiyaY,ChoiG(2009)Genome-wideanalysisofgenestargetedbyPHYTOCHROMEINTERACTINGFACTOR3-LIKE5duringseedgerminationinArabidopsis.
PlantCell21:403-419PerrucE,CharpenteauM,RamirezBC,JauneauA,GalaudJP,RanjeveR,RantyB(2004)Anovelcalmodulin-bindingproteinfunctionsasanegativeregulatorofosmoticstresstoleranceinArabidopsisthalianaseedlings.
PlantJ38:410-420ReedJW,NagataniA,ElichTD,FaganM,ChoryJ(1994)PhytochromeAandphytochromeBhaveoverlappingbutdistinctfunctionsinArabidopsisdevelopment.
PlantPhysiol104:1139–1149ReedJW,NagpalP,PooleDS,FuruyaM,ChoryJ(1993)Mutationsinthegeneforthered/far-redlightreceptorphytochromeBaltercellelongationandhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
31physiologicalresponsesthroughoutArabidopsisdevelopment.
PlantCell5:147-157RushtonPJ,SomssichIE,RinglerP,ShenQJ(2010)WRKYtranscriptionfactors.
TrendsPlantSci15:247-258ShenH,MoonJ,HuqE(2005)PIF1isregulatedbylight-mediateddegradationthroughtheubiquitin-26SproteasomepathwaytooptimizeseedlingphotomorphogenesisinArabidopsis.
PlantJ44:1023–1035ShenH,ZhuL,CastillonA,MajeeM,DownieB,HuqE(2008)Light-inducedphosphorylationanddegradationoftheneagtiveregulatorPHYTOCHROME-INTERACTINGFACTOR1fromArabidopsisdependuponitsdirectphysicalinteractionwithphotoactivatedphytochrome.
PlantCell20:1586-1602ShinJ,KimK,KangH,ZulfugarovIS,BaeG,LeeCH,LeeD,ChoiG(2009)Phytochromespromoteseedlinglightresponsesbyinhibitingfournegatively-actingphytochrome-interactingfactors.
ProcNatlAcadSciUSA106:7660-7665StewartJL,MaloofJN,NemhauserJL(2011)PIFgenesmediatetheeffectofsucroseonseedlinggrowthdynamics.
PLoSOne5:e19894TangW,WangW,ChenD,JiQ,JingY,WangH,LinR(2012)Transposase-derivedproteinsFHY3/FAR1interactwithPHYTOCHROME-INTERACTINGFACTOR1toregulatechlorophyllbiosynthesisbymodulatingHEMB1duringdeetiolationinArabidopsis.
PlantCell24:1984-2000VonArnimA,DengXW(1996)Lightcontrolofseedlingdevelopment.
AnnuRevPlantPhysiolPlantMolBiol47:215–243WalterM,ChabanC,SchutzeK,BatisticO,WeckermannK,NakeC,BlazevicD,GrefenC,SchumacherK,OeckingC,HarterK,KudlaJ(2004)Visualizationofproteininteractionsinlivingplantcellsusingbimolecularfluorescencecomplementation.
PlantJ40:428-438WangA,GarciaD,ZhangH,FengK,ChaudhuryA,BergerF,PeacockWJ,DennisES,LuoM(2010)TheVQmotifproteinIKU1regulatesendospermhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
32growthandseedsizeinArabidopsis.
PlantJ63:670-679WangH,DengXW(2002)ArabidopsisFHY3definesakeyphytochromeAsignalingcomponentdirectlyinteractingwithitshomologouspartnerFAR1.
EMBOJ21:1339-1349WeiN,DengXW(1996)TheroleoftheCOP/DET/FUSgenesinlightcontrolofArabidopsisseedlingdevelopment.
PlantPhysiol112:871–878XieYD,LiW,GuoD,DongJ,ZhangQ,FuY,RenD,PengM,XiaY(2010)TheArabidopsisgeneSIGMAFACTOR-BINDINGPROTEIN1playsaroleinthesalicylate-andjasmonate-mediateddefenseresponses.
PlantCellEnviron33:828-839YadavV,MallappaC,GangappaSN,BhatiaS,ChattopadhyayS(2005)Abasichelix-loop-helixfactorinArabidopsis,MYC2,actsasarepressorofbluelight-mediatedphotomorphogenicgrowth.
PlantCell17:1953-1966https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
33FigureLegendsFigure1.
VQproteinspossesstranscriptionalactivity.
(A)Diagramsofconstructsusedinthetransientexpressionassay.
(B,C)Relativeluciferasereporter(LUC)activitybyvariousVQeffectors.
(D)RelativeLUCreporteractivitybyVQ29anditspointmutants.
For(B-D),theeffectors,LUCreporter,andGUSinternalcontrolwereco-transformedintoArabidopsisprotoplasts.
Datadenotethemean±SDofthreebiologicalreplicates.
Asterisksin(BandC)indicatesignificantdifferencefromtheemptyvectoratP<0.
05(singleasterisk)or0.
01(doubleasterisks)usingStudent'st-test.
Asterisksin(D)indicatesignificantdifferencefromthewild-typeVQ29atP<0.
01(doubleasterisks)usingStudent'st-test.
Figure2.
PhenotypicanalysisoftheVQ29loss-of-functionmutantandoverexpressionplants.
(A)DiagramofVQ29genestructureandthepositionoftheT-DNAinsertion.
Blackrectangle,exonofVQ29.
Triangle,T-DNAinsertion.
(B)DetectionofVQ29transcriptlevelinthewildtypeandvq29-1mutantbyreal-timeRT-PCR.
Seedlingsweregrowninwhitelightconditions(10molm-2s-1)for5d.
Datarepresentthemean±SDofbiologicaltriplicates.
(C)ImmunoblotanalysisoftwoPro35S:Myc-VQ29overexpressiontransgeniclines(VQ29-OE,line#10and#14)andthewildtypeusinganti-MYCantibody.
Immunoblottingagainstthetubulinantibody(anti-TUB)servedasaloadingcontrol.
Seedlingsweregrowninlowwhitelight(10molm-2s-1)for5dbeforeharvesting.
(D)Photomorphogenicresponsesunderfar-red(12molm-2s-1),lowwhitelight(10molm-2s-1)orindarkness.
Seedlingsweregrowninvariousconditionsfor5d.
Barsindicate5mm.
(E)Quantificationofhypocotyllengthoftheseedlingsshownin(D).
Dataaremean±SDofatleast20seedlings.
AsterisksindicatesignificantdifferencefromthewildtypeatP<0.
05(singleasterisk)or0.
01(doubleasterisks)usingStudent'st-test.
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
34(F)ExpressionofPIL1andXTR7byqRT-PCR.
Four-day-olddark-grownseedlingsweretransferredtofar-redlightfor12h.
ErrorbarsindicatetheSDofbiologicaltriplicates.
Figure3.
LocalizationofVQ29.
(A)TheVQ29-GFPandPIF1-GFPconstructsandemptyvectorcontrolsweretransformedintoArabidopsisprotoplasts.
Chlorophyllauto-fluorescencewasshowninred.
Representativeimagesareshown.
Bardenotes5m.
(B)GFPfluorescenceinthehypocotylofPro35S:VQ29-GFPtransgenicseedlingcoincideswithDAPIstaining(whichmarksnucleus).
Seedlingsweregrownindarknessfor4d.
Bardenotes50m.
(C)ImmunoblottingassayofVQ29.
ThePro35S:Myc-VQ29(line#10)transgenicplantsweregrownin16hlight/8hdarkconditionsfor3weeks.
Immunoblottingwithantibodiesagainstcytoplasm-localizedfructose-1,6-bisphosphatase(cFBPase)andplasmamembrane-targetedH+-ATPase(Iwataetal.
,2008)servesaspositivecontrols.
Asteriskindicatesthespecificband.
T,totalprotein;PM,plasmamembrane;C,cytoplasm;N,nucleus.
Figure4.
ExpressionpatternofVQ29.
(A)RT-PCRofVQ29invarioustissues.
Amplifiedactinservedasloadingcontrols.
(B)GUSstaininginvarioustissuesofProVQ29:GUStransgenicplants.
(C)QuantitativeRT-PCRanalysisofVQ29inthephotoreceptormutantsundervariouslightconditionsanddarkness.
Seedlingsweregrownintheindicatedconditionsfor5d.
Dataindicate±SDofthreebiologicaltriplicates.
Figure5.
VQ29physicallyinteractswithPIF1.
(A)Yeasttwo-hybridanalysisoftheinteractionbetweenLexA-VQ29(fusedwiththeLexADNA-bindingdomain)andvariousproteinstaggedwiththeB42activationdomain(AD).
"-"meansemptyADvector.
(B)Relativebeta-galactocidaseactivityofinteractionsbetweenLexA-VQ29withhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
35AD-taggedproteinsshownin(A).
Dataindicate±SDofsixindividualyeastcolonies.
AsterisksindicatesignificantdifferencefromthecombinationofLexA-VQ29andADatP<0.
01usingStudent'st-test.
(C)BiFCassayshowingtheinteractionbetweenPIF1-YFPCandYFPN-VQ29orYFPN-VQ29(V70D,Q71L).
Chlorophyllauto-fluorescencewasshowninred.
Bardenotes5m.
(D)Co-immunoprecipitationassaybetweenVQ29andPIF1.
Pro35S:VQ29-GFPandPro35S:VQ29-GFP/Pro35S:TAP-PIF1seedlingsweregrownindarknessfor5d.
Afterprecipitationwithanti-MYCantibody(TAP-PIF1containsMYCtag),proteinswereimmunoblottedwithanti-MYCoranti-GFPantibodies.
IP,immunoprecipitation.
Figure6.
AnalysisofdoublemutantsandoverexpressionplantsofVQ29andPIF1.
(A)PhotomorphogenicphenotypeoftheColwildtype,vq29,pif1,andvq29pif1mutantsindarkness,orunderfar-red(12molm-2s-1)orlowwhitelight(10molm-2s-1)conditions.
Barsdenote5mm.
(B)Hypocotyllengthofseedlingsshownin(A).
Seedlingsweregrownintheindicatedlightconditionsfor5d.
(C)Hypocotyllengthofseedlingsgrownunderfar-red-darkphotocycles.
Three-day-olddark-grownseedlingsweretransferredtofar-red-dark(FR12h/D12h)conditionatthebeginningoflighttreatmentandgrownforanadditional2d.
(D)HypocotyllengthofPro35S:Myc-VQ29(VQ29-OE,line#10)andPro35S:TAP-PIF1(PIF1-OE)plantsandtheircorrespondingdoubletransgeniclineunderdarknessorlowlightconditions(10molm-2s-1).
Seedlingsweregrownintheindicatedlightconditionsfor5d.
In(B-D),dataaremean±SDofatleast20seedlings.
AsterisksindicatesignificantdifferencefromthewildtypeatP<0.
05(single)or0.
01(double)usingStudent'st-test.
Figure7.
VQ29andPIF1co-regulatedownstreamgeneexpression.
(A)RelativeexpressionofPIL1andXTR7byqRT-PCR.
Seedlingsweregrowninhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
36darknessfor4dandthenkeptindarknessortransferredtofar-redlight(12molm-2s-1)foranadditional1h.
Errorbarsindicate±SDoftriplicates.
(B)Relativeluciferase(LUC)reporteractivityinArabidopsisprotoplastsco-transformedwiththeeffectorconstructs.
LUCgenewasunderthecontrolofeitherPIL1orXTR7promoter.
Datarepresentthemean±SDoftriplicates.
AsterisksindicatesignificantdifferenceatP<0.
01usingStudent'st-test.
(C)RelativeexpressionofPIL1andXTR7inVQ29and/orPIF1overexpressionplantsbyqRT-PCR.
Seedlingsweregrownindarknessfor5d.
Errorbarsindicate±SDoftriplicates.
(D)DiagramofthegenomicstructuresofPIL1andXTR7.
RectanglesdenoteexonsandtrianglesrepresentG-boxmotifs.
AmpliconsusedintheChIPassayareunderlined.
bp,basepairs.
(E,F)ChIP-qPCRassayshowingtherelativeenrichmentoffragmentscorrespondingtothepromotersorcodingregionsofPIL1,XTR7,andtheUBQ1control.
DNAwasprecipitatedwithanti-GFPantibodyinPro35S:VQ29-GFP(E),oranti-MYCantibodyinPro35S:TAP-PIF1(F)transgenicplants,respectively.
Datarepresentthemean±SDoftriplicates.
Figure8.
AproposedmodelofVQ29inregulatingseedlingde-etiolation.
Far-redlight,whichisperceivedbyphytochromes,repressesPIF1andVQ29levels.
VQ29interactswithPIF1andactsasatranscriptionalco-regulatortopromotePIF1'sactivationactivityoncellelongation-relatedgenes,leadingtothesuppressionofseedlingde-etiolation.
Meanwhile,VQ29mightregulatedownstreamgeneexpressionindependentofPIF1.
Arrowsdenotepositiveeffect,andbarsindicatenegativerole.
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
DownloadedonCopyright(c)202DownloadedonCopyright(c)202DownloadedonCopyright(c)202DownloadedonCopyright(c)202DownloadedonCopyright(c)202DownloadedonCopyright(c)202DownloadedonCopyright(c)202https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
ac.
cnKeyLaboratoryofPhotobiology,InstituteofBotany,ChineseAcademyofSciences,Beijing100093,ChinaOne-sentencesummary:AnewtranscriptionregulatorVQ29interactswiththetranscriptionfactorPIF1tomodulatehypocotylcellgrowthinresponsetolight.
PlantPhysiologyPreview.
PublishedonFebruary25,2014,asDOI:10.
1104/pp.
113.
234492Copyright2014bytheAmericanSocietyofPlantBiologistshttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
2ArabidopsisVQ-motif-containingProtein29RepressesSeedlingDe-etiolationbyInteractingwithPIF1YunliangLi,a,bYanjunJing,aJunjiaoLi,a,cGangXu,a,bRongchengLina,1aKeyLaboratoryofPhotobiology,InstituteofBotany,theChineseAcademyofSciences,Beijing100093,ChinabUniversityoftheChineseAcademyofSciences,Beijing100049,ChinacCollegeofLifeSciences,CapitalNormalUniversity,Beijing100048,China1Correspondingauthor,e-mail:rclin@ibcas.
ac.
cnTheauthorresponsiblefordistributionofmaterialintegraltothefindingspresentedinthisarticleinaccordancewiththepolicydescribedintheinstructionsforauthors(www.
plantphysiol.
org)isRongchengLin(rclin@ibcas.
ac.
cn).
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
3Footnotes:ThisworkwassupportedbygrantsfromtheNationalNaturalScienceFoundationofChina(31170221,31325002),andtheMinistryofAgricultureofChina(2011ZX08009-003)toR.
L.
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
4ABSTRACTSeedlingde-etiolation,acriticalprocessinearlyplantdevelopment,isregulatedbyanintricatetranscriptionalnetwork.
Here,weidentifiedVQmotif-containingprotein29(VQ29)asanovelregulatorofthephotomorphogenicresponseinArabidopsisthaliana.
Weshowedthat29ofthe34VQproteinspresentinArabidopsisexhibittranscriptionalactivityinplantcells,andthatmutationsintheVQmotifaffectthetranscriptionalactivityofVQ29.
WethenfunctionallycharacterizedVQ29andshowedthatthehypocotylgrowthofplantsoverexpressingVQ29ishyposensitivetofar-redandlowintensitywhitelight,whereasavq29loss-of-functionmutantexhibitsdecreasedhypocotylelongationunderlowintensityoffar-redlightorwhitelight.
Consistentwiththis,VQ29expressionisrepressedbylightinaphytochrome-dependentmanner.
Intriguingly,ouryeasttwo-hybrid,bimolecularfluorescencecomplementationandco-immunoprecipitationassaysshowedthatVQ29physicallyinteractswithPHYTOCHROME-INTERACTINGFACTOR1(PIF1).
WethenshowedthatVQ29andPIF1directlybindtothepromoterofacellelongation-relatedgene,XYLOGLUCANENDOTRANSGLYCOSYLASE7,andco-activateitsexpression.
Furthermore,thevq29pif1doublemutanthasshorterhypocotylsthaneitherofthecorrespondingsinglemutants.
Therefore,ourstudyrevealsthatVQ29isanegativetranscriptionalregulatoroflight-mediatedinhibitionofhypocotylelongationthatlikelypromotesthetranscriptionalactivityofPIF1duringearlyseedlingdevelopment.
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
5INTRODUCTIONLightisanimportantenvironmentalsignalthataffectsplantgrowthanddevelopmentthroughoutitslifecycle,directingprocessessuchasseedgermination,seedlingde-etiolation,phototropism,circadianrhythms,shadeavoidance,andfloweringtiming.
Dark-grownseedlings,whichadoptadevelopmentalprogramknownasetiolationorskotomorphogenesis,exhibitelongatedhypocotylsandclosedcotyledonswithapicalhooks.
Uponlightirradiation,seedlingsundergode-etiolationorphotomorphogenesis,whichslowshypocotylgrowthandcausescotyledonstoexpandandchloroplastsandchlorophyllstodevelop(vonArnimandDeng,1996).
Intensiveresearchhasrevealedthemainsignalingpathwaygoverningphotomorphogenesis(Chenetal.
,2004,LauandDeng,2010,Arsovskietal.
,2012).
Toinitiatethelightresponses,plantsrelyonasetofphotoreceptors,includingthered/far-redlight-absorbingphytochromes(phys)andtheblue/UV-Alight-absorbingcryptochromes(crys).
Activationofphotoreceptorstransmitssignalstokeydownstreamnegativefactors,suchasCOP1(CONSTITUTIVEPHOTOMORPHOGENIC1)andmembersofthePIF(PHYTOCHROMEINTERACTINGFACTOR)proteinfamily.
COP1isaRING-typeE3ubiquitinligasethattargetsphotomorphogenesis-promotingfactors,suchasHY5(ELONGATEDHYPOCOTYL5)andHFR1(LONGHYPOCOTYLINFAR-RED1),for26Sproteasome-mediateddegradation,whichdesensitizesthelightpathwayinitiatedbybothphysandcrys(WeiandDeng,1996,LauandDeng,2012).
PIFsencodeagroupofbasichelix-loop-helix(bHLH)transcriptionfactors(TFs)thatarephosphorylatedanddegradedinaphy-dependentmannerinlight.
PIFproteins(includingPIF1,3,4,and5)playredundantrolesindirectlyregulatinggeneexpressionandrepressingphotomorphogenicresponses(Leivaretal.
,2008,Shinetal.
,2009,LeivarandQuail,2010).
GeneticandmolecularstudiesinArabidopsisthalianahaveidentifiedanotherseriesofsignalingcomponentsthatisinvolvedineithernegativelyorpositivelyregulatingthelightpathway(Jiaoetal.
,2007).
Notably,manyoftheidentifiedhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
6effectorsarenuclear-localizedTFs,suchasFHY3andFAR1(transposase-derivedTFs),bZIP16(bZIPTFs),HFR1andMYC2(bHLHTFs),LAF1(aMYBTF),andSTH2andLZF1(B-box-containingTFs)(Hudsonetal.
,1999,WangandDeng,2002,Ballesterosetal.
,2001,Hsiehetal.
,2012,Dattaetal.
,2007,Changetal.
,2011,Yadavetal.
,2005).
Moreover,genesencodingF-boxproteins(suchasEID1andAFR),kinases(e.
g.
,NDPK2),andphosphatases(suchasPP7)werealsoisolatedandshowntomediatelightsignaling(Dieterleetal.
,2001,HarmonandKay,2003,Choietal.
,1999,Mlleretal.
,2003).
However,theregulatorymechanismsunderlyingtheactionsofthesefactorsarenotwellunderstood.
Inaddition,werecentlyshowedthatachromatinremodelingfactor,EPP1(enhancedphotomorphogenesis1)/PICKLE,interactswithHY5tofine-tunelightsignalingbymodulatingH3k27me3levelsoncellelongation-relatedgenes(Jingetal.
,2013).
Eventhoughawiderangeoflightsignalingintermediateshavebeenidentifiedandextensivelystudied,acompletepictureofthelightsignalingpathwayhasyettoemerge.
AgroupofgenesencodingproteinscontainingauniqueandconservedFxxxVQxxTGmotif(termedtheVQmotif)wasrecentlyidentified(Xieetal.
,2010,Chengetal.
,2012).
Theseproteinsinclude34membersinArabidopsisandaredesignatedasVQmotif-containingproteins(VQ).
Thefunctionofafewmembersofthisfamilyhasbeencharacterizedtodate.
Forexample,sigmafactorbindingprotein1(SIB1/VQ23),itsclosehomologSIB2(VQ16),andMAPkinase4substrate1(MKS1/VQ21)arerequiredfortheplantdefenseresponse(Narusakaetal.
,2008,Xieetal.
,2010,Laietal.
,2011,Andereassonetal.
,2005).
Inaddition,IKU1(VQ14)isaregulatorofendospermgrowthandseedsize(Wangetal.
,2010),whileCaMBP25(VQ15)andVQ9functionasnegativeeffectorsofosmoticandsalinitystresstolerance,respectively(Perrucetal.
,2004,Huetal.
,2013).
Itisanticipatedthatthisfamilyrespondstovariousenvironmentalsignalsandplaysdiverserolesinplantdefense,growth,anddevelopment.
However,thefunctionsandregulatorymechanismsofmostVQfamilymembersremainunknown.
Inthisstudy,wedemonstratethattheVQfamilyofproteinslargelypossessestranscriptionalactivities.
Furthermore,wecharacterizedtheregulationandfunctionofhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
7VQ29indetail.
WeshowthatVQ29expressionisdown-regulatedbylight.
OverexpressionofVQ29resultsinhyposensitivityofhypocotylgrowthtofar-redandlowlightconditions,whereasthevq29loss-of-functionmutantexhibitsdecreasedhypocotylelongationunderlowintensityoffar-redandwhitelight,duringseedlingde-etiolation.
WealsodemonstratethatVQ29physicallyinteractswithPIF1andthattheseproteinscooperativelyactivatetheexpressionofdownstreamgenes.
OurstudyidentifiesanovelfactorinphotomorphogenesisandprovidesinsightintotherolesofVQfamilyproteinsinregulatingdiverseplantgrowthanddevelopmentalprocesses.
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
8RESULTSAnalysisofVQgenesfromArabidopsis,rice,andmossPreviousstudiesdocumentedthattheVQgenefamilyisfoundonlyinplantsanditwassystematicallystudiedinArabidopsisthaliana(Xieetal.
,2010,Chengetal.
,2012).
Togaininsightintotheevolutionofthisfamily,wesearchedtheGenBankdatabaseforsequencesoftheVQgenesfromrice(Oryzasativa)andmoss(Physcomitrellapatens),whichrepresentmonocotandlowerplants,respectively.
WhereasthemodeldicotplantArabidopsishas34VQmembers,riceandmosshave39and25,respectively.
Interestingly,ArabidopsisandriceVQproteinsarerelativelysmall,withabout85%and92%oftheproteinscontainingfewerthan300aminoacidresidues,respectively.
Incontrast,68%ofthemossVQproteinsarelongerthan300aminoacids(SupplementalFigureS1A).
WefurtherfoundthatmostVQgenesinhigherplants(30inArabidopsisand37inrice)donotcontainanintron.
However,inmoss,only7VQgenesdonothaveintron,whereas5VQshaveoneintronand13genespossesstwoormoreintrons(SupplementalFigureS1B).
TheseresultssuggestthatVQgenestendtobeintronlessandencoderelativelysmallproteinsinhigherplants.
VQproteinsexhibittranscriptionalactivityToinvestigatewhethertheVQproteinsareinvolvedintranscriptionalregulation,weattemptedtoisolatetheopenreadingframe(ORF)ofeachVQgeneinArabidopsisColumbia(Col)plantsusingreversetranscriptionpolymerasechainreaction(RT-PCR,forVQ2)orPCRforotherintronlessVQgenes.
ThePCRprimersweredesignedaccordingtotheavailablecDNAsequenceinformationorthepredictedsequences.
TheORFsofallVQgeneswereamplifiedandclonedintothepEASYvectorandverifiedbysequencing.
WethensubclonedtheVQgenesin-framewiththeGAL4DNA-bindingdomain(GBD)andunderthecontrolofthecauliflowermosaicvirus(CaMV)35SpromoterinthepSAT-GAL4DBvector(Jingetal.
,2013).
Theconstructwasco-transformedwithaluciferasereportergene(LUC),drivenbythe35Sminimalhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
9promoterandfusedin-frametoaGAL4bindingsequence,intoArabidopsismesophyllprotoplasts(Figure1A).
WefoundthatGBDfusionproteinswithVQ14,VQ5,VQ15,VQ16,VQ9,VQ23,VQ3,VQ24,VQ34,VQ17,VQ32,orVQ30drasticallyactivatedLUCreporterexpressioncomparedwithGBDalone.
TheVQ26,VQ12,VQ18,VQ28,andVQ6fusionspromotedLUCexpressiontoalesserextent(Figure1B).
However,GBDfusionswithVQ19,VQ31,VQ4,VQ8,VQ13,VQ33,VQ11,VQ29,VQ7,VQ2,VQ21,orVQ20,remarkablyrepressedtheexpressionoftheLUCreporter(Figure1C).
WealsoobservedthatGBDfusionswithVQ22,VQ1,VQ27,VQ25,andVQ10didnotaffectthetranscriptionofLUC(Figure1B,C).
ThesedataindicatethatmostmembersoftheVQfamilypossesseithertranscriptionalactivationorrepressionactivitiesinthisheterologousgenereportersystem.
Inthisstudy,wefocusedonVQ29(At4g37710)becauseitisinvolvedinseedlingde-etiolationresponse(seebelowindetail).
MutationintheVQmotifaffectstranscriptionalactivitySinceVQfamilymemberscontainonlytheVQmotifandpossessactivationorrepressionactivity,weaskedwhethertheVQmotifisrequiredfortranscriptionalactivity.
WethenalteredconservedaminoacidsintheVQmotifofVQ29,wheretheveryhydrophobicresiduevaline(V)waschangedintoalesshydrophobicresiduealanine(A)orthehydrophilicresidueasparticacid(D),andthehydrophilicresidueglutamine(Q)wasmutagenizedintothehydrophobicresidueleucine(L).
AsshowninFigure1D,singlemutationofVQ29(V70A)orVQ29(V70D)abolishedtherepressiveactivityofVQ29,andgreatlyactivatedLUCreportergeneexpression,whereasmutationinVQ29(Q71L)didnotaffecttheactivity.
Furthermore,doublemutationsinVQ29(V70D,Q71L)ledtoasignificantinductionofLUCtoalesserextentthanVQ29(V70D)(Figure1D).
Itshouldbenotedthattwopointmutations(V70DandQ71L)astested,didnotaffectthelevelsoftheVQ29protein(SupplementalFigureS2).
Takentogether,theseresultssuggestthattheVQmotifislikelyinvolvedinmediatingthetranscriptionalactivityofVQproteins.
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
10OverexpressionofVQ29reducesthehypocotylgrowthresponseunderfar-redandlowintensityofwhitelightconditionsToelucidatethebiologicalfunctionofVQ29,weobtainedtwoT-DNAinsertionlinesofVQ29,Salk_061586andSalk_061438.
PCRgenotypingandsequencingstudiesrevealedthattheT-DNAisinsertedinthepromoterregion136basepairsupstreamoftheATGstartcodonofVQ29inbothmutants.
Thesemutantsthusrepresentthesameallele(Figure2A).
TheyhadbeenbackcrossedfivetimeswiththeColwildtypeandthusthepotentialbackgroundmutationswerelargelyeliminated.
QuantitativeRT-PCRanalysisshowedthattheVQ29transcriptswerebarelydetectableinthemutant(hereafterreferredtoasvq29-1),suggestingthatitisanullallele(Figure2B).
Wealsogeneratedtransgenicplantsover-expressingVQ29,fusedwithaMYCtaganddrivenbytheCaMV35Spromoter(Pro35S:Myc-VQ29,VQ29-OE).
Overfortytransgeniclineswereobtained,twoofwhich(lines#10and#14)werefurtherstudiedinthefollowingexperiments.
ImmunoblottinganalysisusingtheMYCantibodyshowedthatbothlinesaccumulatedMyc-VQ29fusionprotein,withline#10havingahigherlevelofexpressionthanline#14(Figure2C).
Wethentestedthehypocotylgrowthresponseofthevq29-1mutantandVQ29overexpressionlinesundervariouslightconditions.
Thehypocotyllengthofvq29-1wasslightlybutsignificantlyshorterthanthewildtypeunderlowintensityofwhitelight(lessthan20molm-2s-1)orlowintensityoffar-redlight(lessthan12molm-2s-1),butwasindistinguishablefromthatofthewildtypeinredorbluelightconditionswithmultiplefluenceratestested(Figure2D,2E,SupplementalFigureS3).
However,thehypocotylsofVQ29-OEtransgenicseedlingsweresignificantlylongerthanthoseofwild-typeplantsunderfar-redconditions,withline#10exhibitingthestrongerphenotype,consistentwithitshigherVQ29level(Figure2C-E,SupplementalFigureS3).
Thehypocotylsofoverexpressionline#10werealsolongerthanthewildtypeunderlowwhitelightconditions,butnotindarknessorinredorbluelightconditions(Figure2E,SupplementalFigureS3).
Ithasbeendocumentedthatsucrosepromotesseedlinggrowth(Stewartetal.
,2011).
EvenwithoutsucrosesupplementintheMSmedia,vq29-1exhibitedreducedhypocotylelongationunderlowwhitelightandhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
11differentintensitiesoffar-redlight,andthehypocotyllengthofVQ29-OEplants(line#10)waslongerthanthewildtypeundertheseconditions(SupplementalFigureS4).
Moreover,theexpressionoftwolight-responsivegenesinvolvedincellelongation,PHYTOCHROMEINTERACTINGFACTOR3-LIKE1(PIL1)andXYLOGLUCANENDOTRANSGLYCOSYLASE7(XTR7),wasincreasedintheVQ29-OEplantsunderfar-redlightcondition(Figure2F).
Takentogether,theseresultsdemonstratethatVQ29isanegativeregulatorofseedlingde-etiolationunderfar-redandlowwhitelightconditions.
SubcellularlocalizationofVQ29TodeterminethesubcellularlocalizationofVQ29,wefirstfusedVQ29withgreenfluorescenceprotein(GFP)andtransientlyexpressedthisconstructinArabidopsisprotoplasts.
TheVQ29-GFPfusionproteinwasdetectedwithmultiplebandsusingtheGFPantibody,likelyduetopartialdegradationofthefusionproteininvitro(SupplementalFigureS2).
ConfocalmicroscopyrevealedthattheVQ29-GFPfusionwaslikelylocalizedtothenucleus,cytoplasmandplasmamembrane(Figure3A).
Furthermore,wegeneratedstabletransgenicplantsexpressingatranslationalfusionofVQ29withGFPunderthecontroloftheCaMV35Spromoter(Pro35S:VQ29-GFP).
GFPfluorescencewasdistributedmainlyinthenucleusofhypocotylcellsfromthesetransgenicplants(Figure3B).
TosubstantiatethelocalizationofVQ29,weisolatedproteinfractionsfromnucleus,cytoplasmandplasmamembraneofPro35S:Myc-VQ29(line#10)transgenicplants.
AsshowninFigure3C,Myc-VQ29fusionproteinwasonlydetectedincellfractionsisolatedfromthenucleus,butnotfromplasmamembraneorcytosol.
ItslocalizationinthecytoplasmandplasmamembraneintheprotoplastsinFigure3Amightbecausedbyover-expressionofVQ29-GFPintheprotoplasts.
ExpressionpatternofVQ29ToexamineVQ29expressionindifferenttissues,weanalyzeditstranscriptlevelsusingRT-PCR.
RelativelyhighlevelsofVQ29transcriptwereobservedinthestem,https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
12whereasexpressionwaslowintheroot,rosetteleaf,flower,andsilique(Figure4A).
TofurthervisualizetheexpressionpatternofVQ29,wegeneratedtransgenicplantsexpressingtheβ–glucuronidase(GUS)reportergeneunderthecontroloftheVQ29promotersequence(2.
0kbupstreamoftheATGtranslationalstartcodon).
HistochemicalstainingshowedthattheGUSreportergenewasexpressedintheradicle,hypocotyl,stem,leafvein,flower,andsiliquebase,indicatingthatVQ29mayfunctioninthesetissues(Figure4B).
WethenaskedwhetherVQ29isregulatedbylight.
phyA,phyB,andcry1photoreceptormutants,alongwiththeColwildtype,weregrowninfar-red,red,andbluelightconditions,respectively,for5d.
AsshowninFigure4C,quantitativeRT-PCRassaysshowedthatVQ29expressionwasrepressedbyvariouslighttreatments.
Furthermore,theVQ29transcriptlevelswereup-regulatedinthephyA-211andphyB-9mutantsunderfar-redandredlightconditionscomparedwiththewildtype,respectively.
NopronounceddifferenceinVQ29expressionwasfoundbetweenthecry1mutantandthewildtype.
Surprisingly,inthedark,VQ29expressionwasincreasedinphyBmutant,butwasnotaffectedbyphyAandcry1mutations,suggestingthatphyBplaysaroleinregulatingVQ29intheetiolatedseedlings(Figure4C).
TheseobservationssuggestthatlightinhibitsVQ29expressioninaphytochrome-dependentmanner,consistentwithitsroleinregulatingphotomorphogenesisunderfar-redandlowintensitylightconditions.
However,animmunoblottingassayusingtheMYCantibodyandtheVQ29-OEplantsshowedthattheproteinlevelofVQ29wasnotregulatedbylight(SupplementalFigureS5).
VQ29physicallyinteractswithPIF1PreviousstudiesreportedthatVQproteinscouldinteractwithvariousWRKYTFs(Huetal.
,2013,Laietal.
,2011,Wangetal.
,2010).
ThefindingsthatVQ29possessestranscriptionalactivityandisinvolvedinregulatingphotomorphogenesispromptedustohypothesizethatVQ29activitymightdependonitsinteractionwithotherTF(s).
SincePIFproteins,includingPIF1,3,4and5,arebHLH-typeTFsthatplayacriticalroleinrepressingphotomorphogenesis(Leivaretal.
,2008,Shinetal.
,https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
132009),wetestedtheirpossibleinteractionwithVQ29inayeasttwo-hybridsystem.
VQ29wasfusedwiththeLexADNA-bindingdomain(LexA-VQ29)andvariousPIFproteinswereligatedwiththeactivationdomainofB42(AD-PIFs).
Co-expressionofLexA-VQ29withAD-PIF1orAD-PIF3causedstrongactivationoftheLacZreporter(Figure5A,B),indicatingthatVQ29interactswithPIF1andPIF3inyeastcells.
However,onlymildinteractionwasdetectedbetweenVQ29andPIF5.
Asnegativecontrols,AD-SIG1(plastid-locatedSIGMAFACTOR1)orADalonefailedtointeractwithLexA-VQ29(Figure5A,B).
Inthisstudy,therelationshipbetweenVQ29andPIF1wasfurtheranalyzed.
TosubstantiatetheinteractionbetweenVQ29andPIF1inplantcells,weperformedabimolecularfluorescencecomplementation(BiFC)assayinwhichwetransientlyco-expressedtheN-terminusofyellowfluorescenceproteinfusedtoVQ29(YFPN-VQ29)andPIF1fusedtotheC-terminusofYFP(PIF1-YFPC)inArabidopsisprotoplasts(Walteretal.
,2004).
Co-expressionofYFPN-VQ29andPIF1-YFPCreconstitutedafunctionalYFPinthenucleus,whereasco-expressionwitheithercontrolvectorfailedtogenerateYFPfluorescence(Figure5C).
TheinvivointeractionbetweenVQ29andPIF1wasfurtherconfirmedbyco-immunoprecipitationassay.
ThereforewegenerateddoubletransgenicplantsPro35S:VQ29-GFP/Pro35S:TAP-PIF1bycrossingtheirsingletransgenicplant.
TAP-PIF1(usingMYCantibody)wasabletopulldownVQ29-GFP(detectedbyGFPantibody)inthedoubletransgenicseedlings(Figure5D).
Inaddition,twopointmutationsintheVQmotifofVQ29(V70D,Q71L)didnotaffectitsinteractionwithPIF1(Figure5C).
ThesedatatogetherindicatethatVQ29indeedinteractswithPIF1andthattheVQmotifislikelynotrequiredformediatingtheinteraction.
VQ29andPIF1co-regulateseedlingde-etiolationThephysicalinteractionbetweenVQ29andPIF1ledustoinvestigatewhetherandhowtheseproteinsfunctiontogetherinthelightsignalingpathwayinArabidopsis.
Tothisend,wegeneratedthevq29pif1doublemutantbycrossingvq29-1andpif1-2mutants,andVQ29-OEPIF1-OEdoubletransgenicplantsbycrossinghttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
14Pro35S:Myc-VQ29(line#10)andPro35S:TAP-PIF1.
Weexaminedthehypocotylelongationphenotypesofthedoublehomozygouslines,theirsinglemutantortransgenicparentplants,andthewildtype.
Thehypocotyllengthofthevq29pif1doublemutantwasshorterthanthatofthesinglemutantsandColwild-typeseedlingsunderbothfar-redandlowlightconditions(Figure6A,B,SupplementalFigureS3).
Furthermore,thevq29pif1doublemutantwaspronouncedlyshorterthanthesinglemutantsunderfar-red/darkcycles,orunderfar-redlightconditionswithoutsucrosesupplementinthemedia(Figure6C,SupplementalFigureS4B).
Bycontrast,theVQ29-OEPIF1-OEdoubletransgenicplantsdisplayedmuchlongerhypocotylsthantheirparentsingleoverexpressionlinesandthewild-typeplantsunderlowlightconditions(Figure6D).
ThesedatatogetherindicatethatVQ29andPIF1additivelyrepressphotomorphogenesis.
SincePIF1isalsoinvolvedinregulatingphytochrome-mediatedseedgerminationandseedlinggrowthduringde-etiolation(Ohetal.
,2004;Leivaretal.
,2008;Huqetal.
,2004,),wenexttestedwhethermutationoroverexpressionofVQ29havetheseresponses.
Aspreviouslyreported,pif1mutantdisplayedhighgerminationrateafter5minoffar-redlighttreatment(SupplementalFigureS6A).
Seedsofvq29mutantandVQ29-OEdidnotshowanygerminationdifferencecomparedtowildtype,andthegerminationefficiencyofvq29pif1doublemutantwassimilaraspif1(SupplementalFigureS6A).
Inaddition,mutationoroverexpressionofVQ29didnotshowseedlinggreeningphenotype,andvq29-1didnotaffectthedefectsofpif1mutantduringde-etiolation(SupplementalFigureS6B).
TheseobservationsindicatethatVQ29isnotinvolvedinseedgerminationandseedlinggreeningresponseswithPIF1,andthatitsresponsivenessinhypocotylelongationappearstobespecific.
VQ29bindstothepromotersofPIL1andXTR7andregulatestheirexpressionwithPIF1PIF1alsoregulatestheexpressionoflight-responsiveandcellelongation-relatedgenes,suchasPIL1andXTR7(Leivaretal.
,2012).
QuantitativeRT-PCRanalysisrevealedthattheexpressionofPIL1andXTR7wasgreatlydown-regulatedinhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
15darknessinthevq29pif1doublemutantcomparedwithitsparentmutantsandthewildtype(Figure7A).
Inaddition,thetranscriptlevelsofanothertwogenesinvolvedincellelongation,EXTENSIN1(EXT1)andEXT3,weredecreasedinvq29andpif1mutants,andfurtherreducedinthevq29pif1doublemutant(SupplementalFigureS7).
WealsotestedtheexpressionofEXPANSIN8(EXP8)andEXP10,twogenesthatwererepressedbyPIF1(Ohetal.
,2009).
SurprisinglytheEXP10transcriptlevelwasincreasedinvq29pif1doublemutantcomparedthatinpif1(SupplementalFigureS7).
Furthermore,vectorsharboringtheLUCreportergenedriveneitherbythePIL1orXTR7promoterwereconstructedandco-transformedwithVQ29and/orPIF1intoArabidopsisprotoplasts.
AsshowninFigure7B,VQ29itselfdidnotsignificantlypromoteLUCexpressionunderthecontrolofeitherthePIL1orXTR7promoter,whereasPIF1alonestronglyactivatedLUCexpression.
Mostremarkably,co-expressionofPIF1andVQ29increasedtheexpressionlevelsoftheseLUCreportergenes.
Interestingly,amutatedversionofVQ29(V70D)furtheractivatedthelevelsofLUCexpressionwhenco-transformedwithPIF1(Figure7B),suggestingthattheVQdomainofVQ29indeedpossessestranscriptionrepressionfunction.
Consistentwiththeseobservations,theexpressionofPIL1andXTR7wasincreasedinPIF1-OEtransgenicplants,andXTR7'slevelwasfurtheractivatedinVQ29-OEPIF1-OEdoubletransgenicplants(Figure7C).
Therefore,PIF1andVQ29coordinatelyregulatedownstreamgeneexpression,andVQ29likelystimulatestheactivationactivityofPIF1.
ToassesswhethertheinductionofPIL1andXTR7isdirectlyaffectedbyPIF1andVQ29,wecarriedoutchromatinimmunoprecipitationfollowedbyquantitativePCR(ChIP-qPCR)assays.
WhenprecipitatedwithGFPantibodies,fragmentsofthePIL1andXTR7promoterscontainingtheG-box(P),butnottheircodingregions(C),weregreatlyenrichedinextractsfromPro35S:VQ29-GFPtransgenicseedlings,butnotfromColwild-typeseedlings(Figure7D,E).
Inaddition,MYCantibodywasabletopulldownthepromoterfragmentsofXTR7,butnotofPIL1,inextractsfromPro35S:TAP-PIF1transgenicplants(Figure7F).
ThesedataindicatethatVQ29isassociatedwiththepromoterregionsofPIL1andXTR7inplantcells.
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
16PIF1israpidlydegradedafterlighttransition(Shenetal.
,2005;2008).
TotestwhetherVQ29couldaffectthestabilityofPIF1,weperformedanimmunoblottingassayusingtheMYCantibody,andPIF1-OEandPIF1-OEVQ29-OEtransgenicplants.
OurresultshowedthatoverexpressingVQ29didnotaffectthestabilityanddegradationofPIF1(SupplementalFigureS8).
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
17DISCUSSIONVQ29definesanovelrepressorofseedlingde-etiolationInthisstudy,wepresentmultiplelinesofevidencethatVQ29isanoveltranscriptionregulatorthatrepressesseedlingde-etiolationinArabidopsis.
First,hypocotylelongationinplantsoverexpressingVQ29exhibitsreducedsensitivityspecificallytofar-redandlowlightconditions(Figure2).
Itshouldbenotedthatalthoughnoeffectofvq29-1mutantinfar-redlightwasobservedinFigures2Eand6B,astatisticallysignificantshorthypocotylphenotypewasobservedinthismutantinlowfar-redlightintensities(SupplementalFigureS3)orinfar-redlightwithoutsucrose,inmultipleindependentexperiments(SupplementalFigureS4).
Theseoppositephenotypesoftheloss-of-functionmutantandoverexpressionplantsofVQ29suggestthatitscodingproteinisinvolvedinthephytochromesignalingpathway.
Inagreementwiththis,VQ29expressionisdown-regulatedbylightinaphytochrome-dependentmanner(Figure4C).
Second,yeasttwo-hybrid,BiFCandCo-IPassaysshowedthatVQ29isabletophysicallyinteractwithPIF1,akeytranscriptionfactorinthelightsignalingpathway,bothinyeastandinplantcells(Figure5).
Third,thefactsthatthevq29pif1doublemutantorVQ29-OEPIF1-OEdoubletransgenicplantsexhibitedpronouncedphenotypescomparedwiththeircorrespondingsingleparentplantssuggestthatVQ29andPIF1haveadditiveeffect(Figure6,SupplementalFigureS3,S4).
Fourth,VQ29andPIF1functiontogethertoactivatetheexpressionoffourgenesinvolvedinpromotingcellelongation.
Furthermore,thisregulationmightbedirect,asVQ29isassociatedwiththepromotersofPIL1andXTR7invivo(Figure7E).
Thelessstrongphenotypeinthevq29mutantcouldbealternativelycausedbythefunctionalredundancyofVQ29withotherrelatedVQgenes.
FuturestudiesusingdoubleorhighordermutantswithotherVQsshouldfurthertestthispossibility.
PIF1isstabilizedindarknessanddegradedbylight(Shenetal.
,2005,2008).
ItdirectlybindstoDNAviatheG-boxcis-elementinthepromoterofdownstreamgenesandregulatestheirexpression(LeivarandQuail,2010).
Consistently,aG-boxmotifisfoundinthepromoterregionsofbothPIL1andXTR7.
Therefore,weproposehttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
18thatVQ29transcriptaccumulatesindarknessanditscodingproteinactsasatranscriptionco-regulatortopromotetheactivityofPIF1throughphysicalinteraction,leadingtotheactivationofcellelongation-relatedgenes(e.
g.
,PIL1andXTR7),therebyinhibitingseedlingde-etiolation(Figure8).
Infar-redlightconditions,thelevelsofPIF1proteinandVQ29transcriptarereduced,resultingintheinhibitionofcellelongation-relatedgeneexpression,andconsequentlythepromotionofphotomorphogenesis.
Alternatively,VQ29mightfunctionasatranscriptionfactorthatdirectlybindstothepromoterofdownstreamgenesindependentofPIF1.
Furtherstudyisdeservedtoexaminethispossibility.
PIL1(encodesaPIF-regulatedtranscriptionfactor)andXTR7(encodesaxyloglucanendotransglycosylase-relatedprotein)areearlyshademarkergenes(Hornitscheketal.
,2009)thataredirectlyregulatedbyPIF1andVQ29.
VQ29isrequiredforregulatingphotomorphogenesisspecificallyinlowwhitelightandfar-redlight,whichmimictheshadeconditions.
Hence,theinvolvementofVQ29mightfinetunetheresponsivenesstoshade-likeenvironmentbycoordinatingwithPIF1.
Consistently,PIF1alsoplaysarole,althoughmoderate,inshadeavoidanceresponse(Leivaretal.
,2012).
OurChIPexperimentshowedthatVQ29isassociatedwiththepromoterregionofXTR7.
However,nodifferenceintheexpressionofXTR7invq29-1mutantandwildtypewasfound,suggestingthatotherfactor(s)isrequiredforregulatingdownstreamgeneexpressionwithVQ29.
Inagreementwiththisnotion,XTR7mRNAlevelwasgreatlyreducedinthevq29pif1doublemutant.
Inaddition,itisalsopossiblethatotherVQmember(s)playsaroleinregulatingdownstreamgeneexpressionwithVQ29.
AccumulatingstudiessuggestthatmembersoftheVQfamilyplaydiversefunctions,suchasregulatingtheplantdefenseresponse(Narusakaetal.
,2008,Xieetal.
,2010,Laietal.
,2011,Andereassonetal.
,2005),abioticstressresistance(Perrucetal.
,2004,Huetal.
,2013),andseeddevelopment(Wangetal.
,2010).
OurstudyprovidesgeneticandmolecularevidencethatVQ29hasaprominentroleinplantgrowthanddevelopmentinresponsetothelightenvironment,thusextendingthefunctionalityofthisplant-specificproteinfamily.
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
19VQproteinsareinvolvedintranscriptionalregulationVQsaresmallproteinswithashort,uniqueVQmotif.
MolecularstudiesindicatethatVQproteinslikelyinteractwithWRKYtranscriptionfactorsandotherproteins,suchaskinasesandplastidsigmafactors(Laietal.
,2011,Wangetal.
,2010,Chengetal.
,2012,Huetal.
,2013,Andereassonetal.
,2005).
WRKYandPIFareplant-specifictranscriptionfactorsthatplaycrucialbiologicalroles(Rushtonetal.
,2010,LeivarandQuail,2010).
TheinteractionofVQtranscriptionregulatorswithothertranscriptionfactorsappearstofine-tunethetemporalandspatialexpressionpatternsofspecifictargetsduringplantdevelopment,oruponexposuretoparticularbioticandabioticstresses.
OurdatashowedthatmutationsintheVQdomainofVQ29donotaffecttheinteractionwithPIF1(Figure5C).
However,arecentstudyrevealedthattheVQmotifofSIB1isimportantforitsinteractionwithWRKY33(Laietal.
,2011).
Therefore,thereisvariationinwhichdomainsofVQproteinsinteractwithotherproteins.
VQ29possessestranscriptionalrepressionactivityinatransientexpressionsystem.
OurdetailanalysisoftheconservedVandQresiduesintheVQmotifindicatesthatpointmutationsofVQ29alteritstranscriptionalactivity(Figure1D).
Moreover,mutationinVQ29(V70D)furtheractivatesProPIL1:LUCandProXTR7:LUCreporterexpressionmediatedbyPIF1(Figure7B).
TheseobservationsdemonstratethattheVQdomainofVQ29indeedhasrepressivefunctionongeneexpression.
However,VQ29helpsPIF1topromotePIF1-inducedgeneexpression,andtorepresstheexpressionofPIF1-inhibitedgenesduringlight-mediatedearlyseedlinggrowth.
Hence,theactivityofVQ29anditseffectongeneexpressionlikelydependonspecificinteractingfactorandthetargetgenesinagivensignalingprocess.
Similarly,SIB1simulatestheDNA-bindingandtranscriptionalactivityofWRKY33intheplantdefenseresponse(Laietal.
,2011).
Incontrast,recruitmentofVQ9leadstotheinactivationofWRKY8,andtheseproteinsactantagonisticallytomediatethesaltstressresponse(Huetal.
,2013).
OurtransientexpressionassayinprotoplastssuggeststhatthemajorityofVQproteinsexhibiteithertranscriptionalactivationorrepressionactivities(Figure1).
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
20ThefactthatVQ29localizestothenucleusfurthersupportsitsroleinregulatingtranscription.
Furthermore,almostallmembersofthisfamilyarethoughttolocalizetothenucleus,basedonsubcellularproteomicdatabasepredictions(Chengetal.
,2012).
Theidentificationofmoreinteractingproteins,particularlyoftranscriptionfactorsinthenucleus,shouldfurtherelucidatethefunctionsandmechanismsofVQproteinsinplants.
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
21MATERIALSANDMETHODSPlantmaterialsandgrowthconditionsThevq29-1(Salk_061586andSalk_061438),pif1-2,phyA-211,phyB-9andcry1-304mutantsandPro35S:TAP-PIF1transgenicplantswerederivedfromtheArabidopsisthalianaColumbia(Col)ecotype(Chenetal.
,2013,Reedetal.
,1994,1993,Mockleretal.
,1999,Moonetal.
,2008).
vq29-1wasconfirmedbyPCRgenotypingandtheT-DNAinsertionsitewasverifiedbysequencing.
Doublemutantortransgenicplantsweregeneratedbygeneticcrossingandhomozygouslineswereusedintheseexperiments.
SeedlingsweregrownonMurashigeandSkoogmediumcontaining1%sucrose,orwithoutsucroseasindicatedinthetextandSupplementalFigureS4.
Far-redlight(12molm-2s-1),redlight(20molm-2s-1),andbluelight(14molm-2s-1)weresuppliedbylight-emittingdiodelightsources,andlowwhitelight(10molm-2s-1)wassuppliedbycoolwhitefluorescentlamps.
Forfluencerateanalysis,thelightintensitieswereindicatedinthefigures.
DeterminationofHypocotylLength,SeedlingGreeningandSeedGerminationRateForhypocotyllengthmeasurement,seedlingsofdifferentgenotypesweregrownsidebysideonthesameplate,andatleastthreeindependentplateswereusedinallexperiments.
SeedlingswerethenphotographedandtheirhypocotyllengthwasmeasuredbyusingImageJsoftware(http://rsb.
info.
nih.
gov/ij).
Forseedlinggreeningrateanalysis,dark-grownseedlingsweretransferredtocontinuouswhitelightfor2d.
Greeningratewasdeterminedbycountingthepercentageofdark-greencotyledonsfrom50to80seedlingsofeachgenotype.
Forseedgerminationassay,seedsofdifferentgenotypeswereharvestedonthesamedayfromplantsgrowninidenticalconditions.
Seedgerminationwasobservedunderamicroscopeanddeterminedbasedontheappearanceofradicleprotrusion.
Geneexpressionanalysishttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
22PlanttotalRNAwasextractedusinganRNAprepPurePlantKit(Tiangen),andfirststrandcDNAwassynthesizedusingReverseTranscriptase(Invitrogen).
Real-timePCRwascarriedoutusingtheSYBRPremixExTaqKit(Takara)followingthemanufacturer'sinstructions.
Theexpressionlevelswerenormalizedtotheexpressionofaubiquitin(UBQ1)gene.
PrimersarelistedinSupplementalTableS1.
PlasmidconstructionToobtainopenreadingframes(ORFs)ofVQ2,PIF4,andPIF5,firststrandcDNAwasreversetranscribedfromtotalRNAextractedfromColwild-typeseedlingsusingoligo(dT)18primerandhighfidelityPfuDNApolymerase(Invitrogen).
TheproductswereclonedintothepEASYvector(TransGen),resultinginpEASY-VQ2,pEASY-PIF4,andpEASY-PIF5,respectively.
DuetotheabsenceofintronsinallVQgenes(exceptVQ2),theORFswereamplifiedfromgenomicDNAusingPfuandclonedintothepEASYvector,resultinginpEASY-VQs,respectively.
MutationsinVQ29(V70A,V70D,Q71L,V70DQ71L)weregeneratedusingaMutantBESTsite-directedmutagenesiskit(Takara)accordingtothemanufacture'sinstructions.
Appropriaterestrictionenzymesitesweredesignedattheendofeachprimer(SupplementalTableS1).
AllamplifiedORFswerevalidatedbysequencing.
Togenerateconstructsfortheprotoplasttransientexpressionassay,theVQ1fragmentwasreleasedfrompEASY-VQ1digestedwithBamHIandXhoI,andinsertedintotheBglII/SalIsiteofpSAT-GAL4DB(Jingetal.
,2013),toyieldpGAL4DB-VQ1.
TheORFsofVQ4,VQ5,VQ6,VQ7,andVQ18werereleasedfromthecorrespondingMfeI-XhoI-digestedpEASY-VQvectors,respectively,andclonedintotheEcoRI/SalIsitesofpSAT-GAL4DB,givingrisetopGAL4DB-VQ4/5/6/7/18.
TheORFsofVQ24andVQ30werereleasedfrompEASY-VQ24/30digestedwithEcoRIandSalI,andinsertedintotheEcoRI/SalIsitesofpSAT-GAL4DB,generatingpGAL4DB-VQ24andpGAL4DB-VQ30,respectively.
ToobtainpGAL4DB-VQ34,thepEASY-VQ34plasmidwascutwithMfeIandSalI,andtheVQ34ORFwasclonedintotheEcoRI/SalIsitesofpSAT-GAL4DB.
TheORFsofVQ2/3/8/9/10/11/12/13/14/15/16/17/19/20/21/22/23/25/26/27/28/29/31/32/33werehttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
23releasedfromthecorrespondingEcoRI-XhoI-digestedpEASY-VQvectors,respectively,andinsertedintotheEcoRI/SalIsitesofpSAT-GAL4DB,resultinginthecorrespondingpGAL4DB-VQsconstructs.
Toconstructvectorsfortheyeasttwo-hybridassay,thepEASY-VQ29plasmidwascutwithEcoRIandXhoI,andtheVQ29fragmentwasligatedintoEcoRI/XhoI-digestedEG202vector(Clontech),togiverisetopLexA-VQ29.
ThepAD-PIF1andpAD-PIF3plasmidswereconstructedinapreviousstudy(Chenetal.
,2013).
ThepEASY-PIF4andpEASY-PIF5plasmidsweredigestedwithEcoRIandSalI,andthePIF4orPIF5fragmentswereinsertedintotheEcoRI/XhoIsitesoftheJG4-5vector(Clontech),resultinginpAD-PIF4orpAD-PIF5,respectively.
ToprepareconstructsfortheBiFCassay,theVQ29fragmentreleasedfromEcoRI/XhoI–digestedpEASY-VQ29wasinsertedintothepUC-SPYNEvector(Walteretal.
,2004)digestedwithEcoRIandXhoI,togeneratepYFPN-VQ29.
pYFPC-PIF1wasdescribedinapreviousstudy(Chenetal.
,2013).
TostudythesubcellularlocalizationofVQ29,thefragmentreleasedfrompEASY-VQ29usingEcoRIandXhoIwasligatedintotheEcoRI/SalIsitesofthemodifiedpdGNvector(Leeetal.
,2005),togeneratepGFP-VQ29.
ToconstructProVQ29:GUS,afragmentspanningtheregion2-kbupstreamoftheATGstartsiteofVQ29wasamplifiedbyPCRandclonedintopEASY,resultinginpEASY-VQ29p.
ThepEASY-VQ29pplasmidwasthendigestedwithSalIandBamHItoreleasetheVQ29promoterfragment,whichwasthenligatedintothepBI101vectordigestedwiththesameenzymes,togenerateProVQ29:GUS.
ToconstructVQ29overexpressionvectors,theVQ29codingsequencewasreleasedfrompEASY-VQ29usingEcoRIandXhoI,andthenligatedintotheEcoRI-SalIsitesofmodifiedpRI101(Takara),inwhichthreecopiesofMYCtagwereinserted,resultinginPro35S:Myc-VQ29.
TheVQ29ORFwasamplifiedfrompEASY-VQ29byintroducinganNcoIsiteattheN-terminalandaSpeIsiteattheC-terminal,andclonedintopEASYtoproducepEASY-VQ29G.
ThepEASY-VQ29GplasmidwasthendigestedwithNcoIandSpeIandtheVQ29fragmentwasinsertedintopCAMBIA1302(www.
cambia.
org/daisy/cambia/585)cuthttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
24withthesameenzymes,togeneratePro35S:VQ29-GFP.
TogenerateconstructsforthetransientexpressionofPIL1andXTR7,thepromotersequencesofbothgeneswereamplifiedfromColgenomicDNAandligatedintothepEASYvector,resultinginpEASY-PIL1pandpEASY-XTR7p,respectively.
TheseplasmidswerecutwithHindIIIandBamHItoreleasethePIL1andXTR7fragments,whichwereinsertedintotheHindIII/BamHIsitesofthepUC-35sLUCvector(Chenetal.
,2013)toproduceProPIL1:LUCandProXTR7:LUC,respectively.
ThebinaryconstructswereelectroporatedintoAgrobacteriumtumefaciensstrainGV3101andthenintroducedintoColwildtypeplants.
TransgenicplantswereselectedonMSplatesinthepresenceof50mg/Lkanamycinorhygromycin,andhomozygouslineswereusedinthisstudy.
LuciferasetransientexpressionassayTheLUCtransientexpressionassaywascarriedoutaspreviouslydescribed(Jingetal.
,2013).
LUCreporteractivitywasdetectedwithaluminescencekitusingLuciferaseAssaySystem(Promega)andtherelativeactivitywasexpressedastheLUC/GUSratios.
GUShistochemicalassaySeedlingsoftheProVQ29:GUStransgeniclinewereharvestedandincubatedovernightin0.
1Msodiumphosphatebuffercontaining50mMK3Fe(CN)6,50mMK4Fe(CN)6,and1mM5-bromo-4-chloro-3-indolyl-β-D-glucuronideat37°C.
GUSexpressionwasexaminedunderadissectingmicroscopeandimageswerecapturedbyadigitalcamera(Olympus).
GFPproteinlocalizationassayForthetransientassay,VQ29-GFPwastransformedintoArabidopsisprotoplastsandtheprotoplastswereincubatedunderdarknessfor16hbeforeobservation.
ForGFPlocalizationinstabletransgenicplants,ahomozygouslineofPro35S:VQ29-GFPwasused.
TheprotoplastsandtransgenicseedlingsweremountedonaslideandGFPhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
25fluorescencewasvisualizedwithaLeicaTCSSP5confocalmicroscope.
Yeasttwo-hybridassayYeasttwo-hybridanalysiswasperformedaspreviouslydescribed(Tangetal.
,2012).
TransformantsweregrownonSD/-Trp-Ura-HisdropoutplatescontainingX-gal(5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside)forcolordevelopment.
Relativebeta-galactocidaseactivitywasquantifiedaccordingtothemethoddescribedbyYeastProtocolsHandbook(Clontech).
Co-IPandImmunoblottingSeedlingswerehomogenizedinextractionbuffercontaining50mMTris-HCl,pH7.
5,150mMNaCl,10mMMgCl2,0.
1%Tween20,1mMphenylmethylsulfonylfluoride,and1*completeproteaseinhibitorcocktail(Roche).
Theextractswerecentrifugedat14,000*gtwiceat4°Cfor10mineach,andproteinconcentrationwasdeterminedusingtheBradfordassay(Bio-Rad).
Proteinfractionationsfromnucleus,plasmamembraneorcytoplasmwereisolatedaccordingtothemethodsasdescribed(Larssonetal.
,1994,Liuetal.
,2001).
Co-IPassaywascarriedoutaspreviouslydescribed(Tangetal.
,2012).
Proteinswereseparatedon10%SDS-PAGEgelsandtransferredontopolyvinylidenefluoridemembranes.
Theywerethenblottedagainstanti-MYC(Abcam),anti-GFP(Sigma),anti-H+-ATPase(Agrisera),anti-cFBPase(Agrisera),oranti-tubulin(Jingetal.
,2013)antibodies.
Theproteinbandswerevisualizedusingthestandardenhancedchemiluminescencemethod.
BiFCPlasmidscontainingN-andC-terminalYFPfusionswereco-transformedintoArabidopsisprotoplastsaspreviouslydescribed(Walteretal.
,2004).
Theprotoplastswereincubatedunderweaklightfor12-16hbeforeobservation.
YFPfluorescencewascapturedwithaconfocalmicroscope(Leica).
ChIPhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
26TheColwildtypeandPro35S:TAP-PIF1andPro35S:VQ29-GFPtransgenicplantswereusedintheChIPassay,whichwascarriedoutaspreviouslydescribed(Chenetal.
,2013).
Thechromatincomplexeswereincubatedwithanti-MYCoranti-GFPpolyclonalantibodies.
TheprecipitatedDNAfragmentswererecoveredandquantifiedbyquantitativePCRwiththeprimersshowninSupplementalTableS1.
AccessionNumbersSequencedatafromthisarticlecanbefoundintheArabidopsisGenomeInitiativeorGenBank/EMBLdatabasesunderthefollowingaccessionnumbers:VQ29(At4g37710),PIF1(At2g20180),PIF3(At1g09530),PIF4(At2g43010),PIF5(At3g59060),PIL1(At2g46970),XTR7(At4g14130),EXT1(At1g76930),EXT3(At1g21310),EXP8(At2g40610),EXP10(At1g26770),SIG1(At1g08540),ACTIN(At3g18780)andUBQ1(At3g52590),andtheaccessionnumbersofallotherVQgenesarelistedinSupplementalTableS1.
SupplementalDataThefollowingmaterialsareavailableintheonlineversionofthisarticle.
SupplementalFigureS1.
ComparisonofVQgenesinArabidopsis,rice,andmoss.
SupplementalFigureS2.
VQ29proteinlevelinwildtypeandvariouspointmutants.
SupplementalFigureS3.
Fluence-rateresponseunderdifferentlightconditions.
SupplementalFigureS4.
HypocotyllengthofVQ29mutantandoverexpressionplantsgrowninthemediawithoutsucrose.
SupplementalFigureS5.
VQ29proteinlevelduringdark-to-lighttransition.
SupplementalFigureS6.
Phenotypeinseedlinggreeningandseedgermination.
SupplementalFigureS7.
Downstreamgeneexpressioninvq29and/orpif1mutants.
SupplementalFigureS8.
TheeffectofVQ29onPIF1proteinstability.
SupplementalTableS1.
Listofprimersusedinthisstudy.
ACKNOWLEDGMENTShttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
27WethanktheArabidopsisBiologicalResourceCenterforprovidingthevq29mutant.
WearegratefultoDr.
EnamulHuq(UniversityofTexasatAustin)forprovidingthemutantandoverexpressiontransgenicseedsofPIF1,andDr.
TaiWang(InstituteofBotany,ChineseAcademyofSciences)forgiftingplasmamembrane-andcytosol-localizedantibodies.
LITERATURECITEDAndreassonE,JenkinsT,BrodersenP,ThorgrimsenS,PetersenNHT,ZhuS,QiuJL,MicheelsenP,RocherA,PetersenM,NewmanMA,NielsenHB,HirtH,SomssichI,MattssonO,MundyJ(2005)TheMAPkinasesubstrateMKS1isaregulatorofplantdefenseresponses.
EMBOJ24:2579-2589ArsovskiAA,GalstyanA,GusemanJM,NemhauserJL(2012)Photomorphogenesis.
ArabidopsisBook,10:e0147.
doi:10.
1199/tab.
0147BallesterosML,BolleC,LoisLM,MooreJM,Vielle-CalzadaJP,GrossniklausU,ChuaNH(2001)LAF1,aMYBtranscriptionactivatorforphytochromeAsignaling.
GenesDev15:2613–2625ChangCJ,MaloofJN,WuSH(2011)COP1-mediateddegradationofBBX22/LZF1optimizesseedlingdevelopmentinArabidopsis.
PlantPhysiol156:228-239ChenD,XuG,TangW,JingY,JiQ,FeiZ,LinR(2013)AntagonisticbHLH/bZIPtranscriptionfactorsformtranscriptionalmodulesthatintegratelightandreactiveoxygenspeciessignalinginArabidopsis.
PlantCell25:1657-1673ChenM,ChoryJ,FankhauserC(2004)Lightsignaltransductioninhigherplants.
AnnuRevGenet38:87-117ChengY,ZhouY,YangY,ChiYJ,ZhouJ,ChenJY,WangF,FanB,ShiK,ZhouYH,YuJQ,ChenZ(2012)StructuralandfunctionalanalysisofVQmotif-containingproteinsinArabidopsisasinteractingproteinsofWRKYtranscriptionfactors.
PlantPhysiol159:810-825ChoiG,YiH,LeeJ,KwonYK,SohMS,ShinB,LukaZ,HahnTR,SongPS(1999)Phytochromesignalingismediatedthroughnucleosidediphosphatekinasehttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
282.
Nature401:610–613DattaS,HettiarachchiC,JohanssonH,HolmM(2007)SALTTOLERANCEHOMOLOG2,aB-boxproteininArabidopsisthatactivatestranscriptionandpositivelyregulateslight-mediateddevelopment.
PlantCell19:3242-3255DieterleM,ZhouYC,SchaferE,FunkM,KretschT(2001)EID1,anF-boxproteininvolvedinphytochromeA-specificlightsignaling.
GenesDev15:939-944HarmonFG,KaySA(2003)TheFboxproteinAFRisapositveregulatorofphytochromeA-mediatedsignaling.
CurrBiol13:2091-2096HornitschekP,LorrainS,ZoeteV,MichielinO,FankhauserC(2009)Inhibitionoftheshadeavoidanceresponsebyformationofnon-DNAbindingbHLHheterodimers.
EMBOJ28:3893-3902HsiehWP,HsiehHL,WuSH(2012)ArabidopsisbZIP16transcriptionfactorintegrateslightandhormonesignalingpathwaystoregulateearlyseedlingdevelopment.
PlantCell24:3997-4011HuY,ChenL,WangH,ZhangL,WangF,YuD(2013)ArabidopsistranscriptionfactorWRKY8functionsantagonisticallywithitsinteractingpartnerVQ9tomodulatesalinitystresstolerance.
PlantJdoi/10.
1111/tpj.
12159HudsonM,RingliC,BoylanMT,QuailPH(1999)TheFAR1locusencodesanovelnuclearproteinspecifictophytochromeAsignaling.
GenesDev13:2017–2027HuqE,Al-SadyB,HudsonM,KimCH,ApelM,QuailPH(2004)PHYTOCHROME-INTERACTINGFACTOR1isacriticalbHLHregulatorofchlorophyllbiosynthesis.
Science305:1937-1941IwataY,FedoroffNV,KoizumiN(2008)ArabidopsisbZIP60isaproteolysis-activatedtranscriptionfactorinvolvedintheendoplasmicreticulumstressresponse.
PlantCell20:3107–3121.
JiaoY,LauOS,DengXW(2007)Light-regulatedtranscriptionalnetworkinhigherplants.
NatRevGene8:217-230JingY,ZhangD,WangX,TangW,WangW,HuaiJ,XuG,ChenD,LiY,LinRhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
29(2013)Arabidopsischromatin-remodelingfactorPICKLEinteractswithtranscriptionfactorHY5toregulatehypocotylcellelongation.
PlantCell25:242-256LaiZ,LiY,WangF,ChengY,FanB,YuJQ,ChenZ(2011)ArabidopsissigmafactorbindingproteinsareactivatorsoftheWRKY33transcriptionfactorinplantdefense.
PlantCell23:3824-3841LarssonC,SommarinM,WidellS(1994)Isolationofhighlypurifiedplantplasmamembranesandseparationofinside-outandright-side-outvesicles.
MethodsEnzymol228:451-469LauOS,DengXW(2010)Planthormonesignalinglightensup:integratorsoflightandhormones.
CurrOpinPlantBiol13:571-577LauOS,DengXW(2012)ThephotomorphogenicrepressorsCOP1andDET1:20yearslater.
TrendsPlantSci17:584-93LeeJ,TaokaK,YooB,Ben-NissanG,KimD,LucasWJ(2005)Plasmodesmal-associatedproteinkinaseintobaccoandArabidopsisrecognizesasubsetofnon-cell-autonomousproteins.
PlantCell17:2817-2831LeivarP,MonteE,OkaY,LiuT,CarleC,CastillonA,HuqE,QuailPH(2008)Multiplephytochrome-interactingbHLHtranscriptionfactorsrepressprematureseedlingphotomorphogenesisindarkness.
CurrBiol18:1815-1823LeivarP,QuailPH(2010)PIFs:pivotalcomponentsinacellularsignalinghub.
TrendsPlantSci16:19-28LeivarP,TeppermanJM,CohnMM,MontteE,Al-SadyB,EricksonE,QuailPH(2012)DynamicantagonismbetweenphytochromesandPIFfamilybasichelix-loop-helixfactorsinducesselectivereciprocalresponsestolightandshadeinarapidresponsivetranscriptionalnetworkinArabidopsis.
PlantCell24:1398-1419LiuX,CovingtonMF,FankhauserC,ChoryJ,WagnerDR(2001)ELF3encodesacircadianclock–regulatednuclearproteinthatfunctionsinanArabidopsisPHYBsignaltransductionpathway.
PlantCell13:1293-1304MocklerTC,GuoH,YangH,DuongH,Lin,C(1999)Antagonisticactionsofhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
30ArabidopsiscryptochromesandphytochromeBintheregulationoffloralinduction.
Development126:2073–2082MllerSG,KimYS,KunkelT,ChuaNH(2003)PP7isapositiveregulatorofbluelightsignalinginArabidopsis.
PlantCell15:1111–1119MoonJ,ZhuL,ShenH,HuqE(2008)PIF1directlyandindirectlyregulateschlorophyllbiosynthesistooptimizethegreeningprocessinArabidopsis.
ProcNatlAcadSciUSA105:9433-9438MorikawaK,ShiinaT,MurakamiS,ToyoshimaY(2002)Novelnuclear-encodedproteinsinteractingwithaplastidfactor,Sig1,inArabidopsisthaliana.
FEBSLetters514:300-304NarusakaM,KawaiK,izawaN,SekiM,ShinozakiK,SeoS,KobayashiM,ShiraishiT,NarusakaY(2008)GenecodingforSigA-bindingproteinfromArabidopsisappearstobetranscriptionallyup-regulatedbysalicylicacidandNPR1-dependentmechanisms.
JGenPlantPathol74:345-354OhE,KimJ,ParkE,KimJ,KangC,ChoiG(2004)PIL5,aphytochrome-interactingbasichelix-loop-helixprotein,isakeynegativeregulatorofseedgerminationinArabidopsisthaliana.
PlantCell16:3045-3058OhE,KangH,YamaguchiS,ParkJ,LeeD,KamiyaY,ChoiG(2009)Genome-wideanalysisofgenestargetedbyPHYTOCHROMEINTERACTINGFACTOR3-LIKE5duringseedgerminationinArabidopsis.
PlantCell21:403-419PerrucE,CharpenteauM,RamirezBC,JauneauA,GalaudJP,RanjeveR,RantyB(2004)Anovelcalmodulin-bindingproteinfunctionsasanegativeregulatorofosmoticstresstoleranceinArabidopsisthalianaseedlings.
PlantJ38:410-420ReedJW,NagataniA,ElichTD,FaganM,ChoryJ(1994)PhytochromeAandphytochromeBhaveoverlappingbutdistinctfunctionsinArabidopsisdevelopment.
PlantPhysiol104:1139–1149ReedJW,NagpalP,PooleDS,FuruyaM,ChoryJ(1993)Mutationsinthegeneforthered/far-redlightreceptorphytochromeBaltercellelongationandhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
31physiologicalresponsesthroughoutArabidopsisdevelopment.
PlantCell5:147-157RushtonPJ,SomssichIE,RinglerP,ShenQJ(2010)WRKYtranscriptionfactors.
TrendsPlantSci15:247-258ShenH,MoonJ,HuqE(2005)PIF1isregulatedbylight-mediateddegradationthroughtheubiquitin-26SproteasomepathwaytooptimizeseedlingphotomorphogenesisinArabidopsis.
PlantJ44:1023–1035ShenH,ZhuL,CastillonA,MajeeM,DownieB,HuqE(2008)Light-inducedphosphorylationanddegradationoftheneagtiveregulatorPHYTOCHROME-INTERACTINGFACTOR1fromArabidopsisdependuponitsdirectphysicalinteractionwithphotoactivatedphytochrome.
PlantCell20:1586-1602ShinJ,KimK,KangH,ZulfugarovIS,BaeG,LeeCH,LeeD,ChoiG(2009)Phytochromespromoteseedlinglightresponsesbyinhibitingfournegatively-actingphytochrome-interactingfactors.
ProcNatlAcadSciUSA106:7660-7665StewartJL,MaloofJN,NemhauserJL(2011)PIFgenesmediatetheeffectofsucroseonseedlinggrowthdynamics.
PLoSOne5:e19894TangW,WangW,ChenD,JiQ,JingY,WangH,LinR(2012)Transposase-derivedproteinsFHY3/FAR1interactwithPHYTOCHROME-INTERACTINGFACTOR1toregulatechlorophyllbiosynthesisbymodulatingHEMB1duringdeetiolationinArabidopsis.
PlantCell24:1984-2000VonArnimA,DengXW(1996)Lightcontrolofseedlingdevelopment.
AnnuRevPlantPhysiolPlantMolBiol47:215–243WalterM,ChabanC,SchutzeK,BatisticO,WeckermannK,NakeC,BlazevicD,GrefenC,SchumacherK,OeckingC,HarterK,KudlaJ(2004)Visualizationofproteininteractionsinlivingplantcellsusingbimolecularfluorescencecomplementation.
PlantJ40:428-438WangA,GarciaD,ZhangH,FengK,ChaudhuryA,BergerF,PeacockWJ,DennisES,LuoM(2010)TheVQmotifproteinIKU1regulatesendospermhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
32growthandseedsizeinArabidopsis.
PlantJ63:670-679WangH,DengXW(2002)ArabidopsisFHY3definesakeyphytochromeAsignalingcomponentdirectlyinteractingwithitshomologouspartnerFAR1.
EMBOJ21:1339-1349WeiN,DengXW(1996)TheroleoftheCOP/DET/FUSgenesinlightcontrolofArabidopsisseedlingdevelopment.
PlantPhysiol112:871–878XieYD,LiW,GuoD,DongJ,ZhangQ,FuY,RenD,PengM,XiaY(2010)TheArabidopsisgeneSIGMAFACTOR-BINDINGPROTEIN1playsaroleinthesalicylate-andjasmonate-mediateddefenseresponses.
PlantCellEnviron33:828-839YadavV,MallappaC,GangappaSN,BhatiaS,ChattopadhyayS(2005)Abasichelix-loop-helixfactorinArabidopsis,MYC2,actsasarepressorofbluelight-mediatedphotomorphogenicgrowth.
PlantCell17:1953-1966https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
33FigureLegendsFigure1.
VQproteinspossesstranscriptionalactivity.
(A)Diagramsofconstructsusedinthetransientexpressionassay.
(B,C)Relativeluciferasereporter(LUC)activitybyvariousVQeffectors.
(D)RelativeLUCreporteractivitybyVQ29anditspointmutants.
For(B-D),theeffectors,LUCreporter,andGUSinternalcontrolwereco-transformedintoArabidopsisprotoplasts.
Datadenotethemean±SDofthreebiologicalreplicates.
Asterisksin(BandC)indicatesignificantdifferencefromtheemptyvectoratP<0.
05(singleasterisk)or0.
01(doubleasterisks)usingStudent'st-test.
Asterisksin(D)indicatesignificantdifferencefromthewild-typeVQ29atP<0.
01(doubleasterisks)usingStudent'st-test.
Figure2.
PhenotypicanalysisoftheVQ29loss-of-functionmutantandoverexpressionplants.
(A)DiagramofVQ29genestructureandthepositionoftheT-DNAinsertion.
Blackrectangle,exonofVQ29.
Triangle,T-DNAinsertion.
(B)DetectionofVQ29transcriptlevelinthewildtypeandvq29-1mutantbyreal-timeRT-PCR.
Seedlingsweregrowninwhitelightconditions(10molm-2s-1)for5d.
Datarepresentthemean±SDofbiologicaltriplicates.
(C)ImmunoblotanalysisoftwoPro35S:Myc-VQ29overexpressiontransgeniclines(VQ29-OE,line#10and#14)andthewildtypeusinganti-MYCantibody.
Immunoblottingagainstthetubulinantibody(anti-TUB)servedasaloadingcontrol.
Seedlingsweregrowninlowwhitelight(10molm-2s-1)for5dbeforeharvesting.
(D)Photomorphogenicresponsesunderfar-red(12molm-2s-1),lowwhitelight(10molm-2s-1)orindarkness.
Seedlingsweregrowninvariousconditionsfor5d.
Barsindicate5mm.
(E)Quantificationofhypocotyllengthoftheseedlingsshownin(D).
Dataaremean±SDofatleast20seedlings.
AsterisksindicatesignificantdifferencefromthewildtypeatP<0.
05(singleasterisk)or0.
01(doubleasterisks)usingStudent'st-test.
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
34(F)ExpressionofPIL1andXTR7byqRT-PCR.
Four-day-olddark-grownseedlingsweretransferredtofar-redlightfor12h.
ErrorbarsindicatetheSDofbiologicaltriplicates.
Figure3.
LocalizationofVQ29.
(A)TheVQ29-GFPandPIF1-GFPconstructsandemptyvectorcontrolsweretransformedintoArabidopsisprotoplasts.
Chlorophyllauto-fluorescencewasshowninred.
Representativeimagesareshown.
Bardenotes5m.
(B)GFPfluorescenceinthehypocotylofPro35S:VQ29-GFPtransgenicseedlingcoincideswithDAPIstaining(whichmarksnucleus).
Seedlingsweregrownindarknessfor4d.
Bardenotes50m.
(C)ImmunoblottingassayofVQ29.
ThePro35S:Myc-VQ29(line#10)transgenicplantsweregrownin16hlight/8hdarkconditionsfor3weeks.
Immunoblottingwithantibodiesagainstcytoplasm-localizedfructose-1,6-bisphosphatase(cFBPase)andplasmamembrane-targetedH+-ATPase(Iwataetal.
,2008)servesaspositivecontrols.
Asteriskindicatesthespecificband.
T,totalprotein;PM,plasmamembrane;C,cytoplasm;N,nucleus.
Figure4.
ExpressionpatternofVQ29.
(A)RT-PCRofVQ29invarioustissues.
Amplifiedactinservedasloadingcontrols.
(B)GUSstaininginvarioustissuesofProVQ29:GUStransgenicplants.
(C)QuantitativeRT-PCRanalysisofVQ29inthephotoreceptormutantsundervariouslightconditionsanddarkness.
Seedlingsweregrownintheindicatedconditionsfor5d.
Dataindicate±SDofthreebiologicaltriplicates.
Figure5.
VQ29physicallyinteractswithPIF1.
(A)Yeasttwo-hybridanalysisoftheinteractionbetweenLexA-VQ29(fusedwiththeLexADNA-bindingdomain)andvariousproteinstaggedwiththeB42activationdomain(AD).
"-"meansemptyADvector.
(B)Relativebeta-galactocidaseactivityofinteractionsbetweenLexA-VQ29withhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
35AD-taggedproteinsshownin(A).
Dataindicate±SDofsixindividualyeastcolonies.
AsterisksindicatesignificantdifferencefromthecombinationofLexA-VQ29andADatP<0.
01usingStudent'st-test.
(C)BiFCassayshowingtheinteractionbetweenPIF1-YFPCandYFPN-VQ29orYFPN-VQ29(V70D,Q71L).
Chlorophyllauto-fluorescencewasshowninred.
Bardenotes5m.
(D)Co-immunoprecipitationassaybetweenVQ29andPIF1.
Pro35S:VQ29-GFPandPro35S:VQ29-GFP/Pro35S:TAP-PIF1seedlingsweregrownindarknessfor5d.
Afterprecipitationwithanti-MYCantibody(TAP-PIF1containsMYCtag),proteinswereimmunoblottedwithanti-MYCoranti-GFPantibodies.
IP,immunoprecipitation.
Figure6.
AnalysisofdoublemutantsandoverexpressionplantsofVQ29andPIF1.
(A)PhotomorphogenicphenotypeoftheColwildtype,vq29,pif1,andvq29pif1mutantsindarkness,orunderfar-red(12molm-2s-1)orlowwhitelight(10molm-2s-1)conditions.
Barsdenote5mm.
(B)Hypocotyllengthofseedlingsshownin(A).
Seedlingsweregrownintheindicatedlightconditionsfor5d.
(C)Hypocotyllengthofseedlingsgrownunderfar-red-darkphotocycles.
Three-day-olddark-grownseedlingsweretransferredtofar-red-dark(FR12h/D12h)conditionatthebeginningoflighttreatmentandgrownforanadditional2d.
(D)HypocotyllengthofPro35S:Myc-VQ29(VQ29-OE,line#10)andPro35S:TAP-PIF1(PIF1-OE)plantsandtheircorrespondingdoubletransgeniclineunderdarknessorlowlightconditions(10molm-2s-1).
Seedlingsweregrownintheindicatedlightconditionsfor5d.
In(B-D),dataaremean±SDofatleast20seedlings.
AsterisksindicatesignificantdifferencefromthewildtypeatP<0.
05(single)or0.
01(double)usingStudent'st-test.
Figure7.
VQ29andPIF1co-regulatedownstreamgeneexpression.
(A)RelativeexpressionofPIL1andXTR7byqRT-PCR.
Seedlingsweregrowninhttps://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
36darknessfor4dandthenkeptindarknessortransferredtofar-redlight(12molm-2s-1)foranadditional1h.
Errorbarsindicate±SDoftriplicates.
(B)Relativeluciferase(LUC)reporteractivityinArabidopsisprotoplastsco-transformedwiththeeffectorconstructs.
LUCgenewasunderthecontrolofeitherPIL1orXTR7promoter.
Datarepresentthemean±SDoftriplicates.
AsterisksindicatesignificantdifferenceatP<0.
01usingStudent'st-test.
(C)RelativeexpressionofPIL1andXTR7inVQ29and/orPIF1overexpressionplantsbyqRT-PCR.
Seedlingsweregrownindarknessfor5d.
Errorbarsindicate±SDoftriplicates.
(D)DiagramofthegenomicstructuresofPIL1andXTR7.
RectanglesdenoteexonsandtrianglesrepresentG-boxmotifs.
AmpliconsusedintheChIPassayareunderlined.
bp,basepairs.
(E,F)ChIP-qPCRassayshowingtherelativeenrichmentoffragmentscorrespondingtothepromotersorcodingregionsofPIL1,XTR7,andtheUBQ1control.
DNAwasprecipitatedwithanti-GFPantibodyinPro35S:VQ29-GFP(E),oranti-MYCantibodyinPro35S:TAP-PIF1(F)transgenicplants,respectively.
Datarepresentthemean±SDoftriplicates.
Figure8.
AproposedmodelofVQ29inregulatingseedlingde-etiolation.
Far-redlight,whichisperceivedbyphytochromes,repressesPIF1andVQ29levels.
VQ29interactswithPIF1andactsasatranscriptionalco-regulatortopromotePIF1'sactivationactivityoncellelongation-relatedgenes,leadingtothesuppressionofseedlingde-etiolation.
Meanwhile,VQ29mightregulatedownstreamgeneexpressionindependentofPIF1.
Arrowsdenotepositiveeffect,andbarsindicatenegativerole.
https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
DownloadedonCopyright(c)202DownloadedonCopyright(c)202DownloadedonCopyright(c)202DownloadedonCopyright(c)202DownloadedonCopyright(c)202DownloadedonCopyright(c)202DownloadedonCopyright(c)202https://plantphysiol.
orgDownloadedonDecember29,2020.
-PublishedbyCopyright(c)2020AmericanSocietyofPlantBiologists.
Allrightsreserved.
- activationcjblaze相关文档
- Sterncjblaze
- tioncjblaze
- contentcjblaze
- deterministiccjblaze
- optimalcjblaze
- RESULTScjblaze
UCloud优刻得,新增1核1G内存AMD快杰云机型,服务器2元/首月,47元/年
UCloud优刻得近日针对全球大促活动进行了一次改版,这次改版更加优惠了,要比之前的优惠价格还要低一些,并且新增了1核心1G内存的快杰云服务器,2元/首年,47元/年,这个价格应该是目前市面上最低最便宜的云服务器产品了,有需要国内外便宜VPS云服务器的朋友可以关注一下。UCloud好不好,UCloud服务器怎么样?UCloud服务器值不值得购买UCloud是优刻得科技股份有限公司旗下拥有的云计算服...
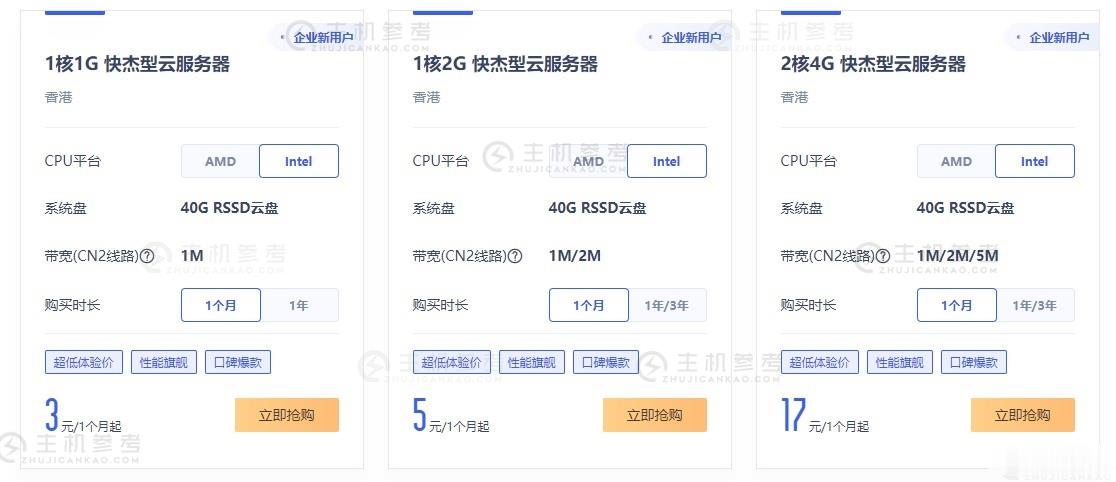
易探云月付18元起,香港/美国/深圳/北京VPS,CN2、BGP等多线路
易探云怎么样?易探云是国内一家云计算服务商家,致力香港服务器、国内外服务器租用及托管等互联网业务,目前主要地区为运作香港BGP、香港CN2、广东、北京、深圳等地区。易探云服务器均选择当下热门线路,比如CN2 GIA、BGP线路、CN2线路等,所有云主机支持月付,并且首月优惠,年付优惠,优惠后香港沙田云服务器/独立ip/香港CN2线路,每月仅18元,188元/年。点击进入:易探云官方网站地址1、香港...
柚子互联(34元),湖北十堰高防, 香港 1核1G 5M
柚子互联官网商家介绍柚子互联(www.19vps.cn)本次给大家带来了盛夏促销活动,本次推出的活动是湖北十堰高防产品,这次老板也人狠话不多丢了一个6.5折优惠券而且还是续费同价,稳撸。喜欢的朋友可以看看下面的活动详情介绍,自从站长这么久以来柚子互联从19年开始算是老商家了。六五折优惠码:6kfUGl07活动截止时间:2021年9月30日客服QQ:207781983本次仅推荐部分套餐,更多套餐可进...

cjblaze为你推荐
-
虚拟主机价格谁知道虚拟主机的价格?域名注册公司域名注册公司是不是要向DNS根服务器交钱?外国虚拟主机为什么淘宝上的 外国的虚拟主机 这么便宜?免费虚拟主机申请谁有1年免费的虚拟主机申请地址吖?国内免费空间免费空间哪个好用国内ip代理找一个好用的国内电信IP代理?虚拟主机申请域名申请以及虚拟主机成都虚拟空间空间服务商那个好虚拟主机软件哪种虚拟机软件好用虚拟主机服务商哪个虚拟主机的服务商比较好?