traits33669.com
33669.com 时间:2021-05-13 阅读:()
RESEARCHARTICLEOpenAccessReclassificationofthetaxonomicstatusofSEMIA3007isolatedinMexicoB-11AMexasRhizobiumleguminosarumbv.
viceaebybioinformatictoolsLucianoTakeshiKishi,CamilaCesárioFernandes,WellingtonPineOmori,JooCarlosCampanharoandElianaGertrudesdeMacedoLemos*AbstractBackground:EvidencebasedongenomicsequencesisextremelyimportanttoconfirmthephylogeneticrelationshipswithintheRhizobiumgroup.
SEMIA3007wasanalyzedwithintheMesorhizobiumgroupstodefinetheunderlyingcausesoftaxonomicidentification.
Wepreviouslyusedbiochemicaltestsandphenotypictaxonomicmethodstoidentifybacteria,whichcanleadtoerroneousclassification.
AnimprovedunderstandingofbacterialstrainssuchastheMesorhizobiumgenuswouldincreaseourknowledgeofclassificationandevolutionofthesespecies.
Results:Inthisstudy,wesequencedthecompletegenomeofSEMIA3007andcompareditwithfiveotherMesorhizobiumandtwoRhizobiumgenomes.
ThegenomesofisolatedSEMIA3007showedseveralorthologswithM.
huakuii,M.
erdmaniiandM.
loti.
WeidentifiedSEMIA3007asaMesorhizobiumbycomparingthe16SrRNAgeneandthecompletegenome.
Conclusion:Ourortholog,16SrRNAgeneandaveragenucleotideidentityvalues(ANI)analysisalldemonstrateSEMIA3007isnotRhizobiumleguminosarumbv.
viceae.
TheresultsofthephylogeneticanalysisclearlyshowSEMIA3007ispartoftheMesorhizobiumgroupandsuggestareclassificationiswarranted.
Keywords:Genomesequencing,Coregenome,Comparativeanalysis,Orthologgenes,PhylogeneticanalysisBackgroundRhizobiaisthecollectivenameofthegeneraRhizobium,SinorhizobiumandMesorhizobium,whicharesoilandrhizospherebacteriaofagronomicimportancebecausetheyformnitrogen-fixingsymbioseswithleguminousplants[1,2].
Thus,rhizobiaareconsideredbio-fertilizersandhavebeenusedasinoculantsforover120years.
Rhizobialgeneticdiversityandtheplant-bacteriamo-lecularinteractionshavebeenwell-studied[3].
ThegrowthrateofMesorhizobiumisintermediatebetweenthegeneraRhizobiumandBradyrhizobiumandisoneofthelargestgenera.
Additionally,theMesorhizobiumgeneraconsistsof24speciesfoundinAsia,Europe,theMediterraneanregionandAfrica[4,5].
Jarvinsetal.
[6]werethefirsttorequestthecreationoftheMesorhizobiumgenusandreclassifiedseveralgeneraidentifiedasRhizobiumintoMesorhizobium.
Thecorrectphylogeneticidentificationofaspeciesrequiresanaccuratetechnicalcharacterization[7,8].
ThetaxonomyofMesorhizobiumrequiresthereclassi-ficationofspeciesbecausethereisaneedforstudiestoavoidclassificationproblems.
Taxonomicinformationprovidesaccesstobasictraitinformationsuchasphysi-ology,epidemiologyandevolutionaryhistory[9].
Thecorrecttaxonomicassignmentofbacterialgenomesisaprimaryandchallengingtask[10–13].
*Correspondence:egerle@fcav.
unesp.
brEqualcontributorsDepartamentodeTecnologia,LaboratóriodeBioquímicadeMicrorganismosePlanta–LBMP,UNESP-UniversidadeEstadualPaulista,FaculdadedeCiênciasAgráriaseVeterinárias,ViadeAcessoProf.
PauloDonatoCastellanes/n,14884-900Jaboticabal,SP,BrazilTheAuthor(s).
2016OpenAccessThisarticleisdistributedunderthetermsoftheCreativeCommonsAttribution4.
0InternationalLicense(http://creativecommons.
org/licenses/by/4.
0/),whichpermitsunrestricteduse,distribution,andreproductioninanymedium,providedyougiveappropriatecredittotheoriginalauthor(s)andthesource,providealinktotheCreativeCommonslicense,andindicateifchangesweremade.
TheCreativeCommonsPublicDomainDedicationwaiver(http://creativecommons.
org/publicdomain/zero/1.
0/)appliestothedatamadeavailableinthisarticle,unlessotherwisestated.
Kishietal.
BMCMicrobiology(2016)16:260DOI10.
1186/s12866-016-0882-5Thepartial16SribosomalRNAgene(16SrRNA)isamolecularmarkerwidelyusedinthetaxonomyofbacteria.
However,thisgenehasnoconsensussequencetocorrectlyclassifymicroorganismsatthespecieslevel[14–16].
Thus,DNA-DNAhybridization(DDH)hasbeenusedasthegoldstandardfordefiningprokaryoticspeciesatthegenomiclevel.
DDHistheonlytaxonomicmethodthatoffersanumericalandrelativelystablespecies.
Therefore,DDHinfluenceshowthecurrentclassificationsystemhasbeenconstructed[17].
DDHisanexpensiveandlaboriousmethodthatisavailableinonlyafewlaboratoriesworldwide,sinceitrequiresthehybridizationofhundredsofstrainsandoftendoesnotresolvethetaxonomicproblems.
How-ever,itisanimportantlimitingfactorforthedescriptionofnewspecies,particularlyincountrieswiththegreatestbiodiversity.
ProkaryoticspeciescontinuetobeagroupofstrainsduetoDNA-DNAre-associationvaluesgreaterthan70%[14,18].
Therecentdevelopmentofsequencingtechnologieshasenabledustocarefullyassessmicrobialcommunitiesbygeneratingmanynucleotidesequencesatlowercosts.
Nextgenerationsequencing(NGS)technologieshaverevolutionizedthefieldofmicrobialecologyandallowsresearcherstodeterminethelevelofdiversitymorecloselyusingin-depthsequencing.
TherearevariousapplicationsusingtheseNGSplatforms,whichrangefromsingle-genetargetedsequencingtowhole-genomesequencingandshotgunmetagenomesequencing[19].
Withtheavailabilityofwholegenomesequences,thegenecontentbasedapproachesappearpromisingininferringthebacterialtaxonomy.
Thecompletegenomesequencingofabacterialgenomeoftenrevealsasubstantialnumberofuniquegenespresentonlyinthatgenomewhichcanbeusedforitstaxonomicclassification[11,12].
TherecentimprovedaccesstovariousnewgenesequencesandthedefinitionofprokaryotespecieshasledtodoubtsregardingthesuitabilityoftheDNA-DNAhybridizationmethod[20].
Thenewproposalsincludetheanalysisofseveralgenesortheentiregenome.
Oneproposedanalysismethodistoanalyzecommongenesbetweentwostrainsanddeterminetheaveragenucleo-tideidentityvalues(ANI).
AnANIvalueexceeding94%correspondsto70%traditionalDNA-DNAhybridization[21,22].
Thisanalysismethodalsoconsidersgeneswitheco-logicalfunctions.
OtherANIvaluessuggestedreplacing70%hybridizationwith95%ANIand69%conservedDNA.
Intheproteincodingportionofthegenome,thesevalueswouldsuggest85%conservedgenes[23].
Themostrecentproposalsrecommend>95–96%ANItodelineatespeciesandwouldreplacethetraditional70%cutoffthresholdusedforDDHsequences[17].
TheaimofthisstudywastoevaluateSEMIA3007isolatedinMexicoasB-11AMexandisclassifiedbyphenotypictaxonomicmethodssuchasRhizobiumlegu-minosarumbv.
viceaebydifferentgroupsofresearchers.
Weusedacombinationofcomplete16SrRNAsequen-cingandcompletegenomeanalysistoreclassifyB-11AasMesorhizobiumsp.
ResultsanddiscussionBacterialgrowthcurveThebacterialgrowthcurveofSEMIA3007isshowninFig.
1.
SEMIA3007grewsimilartothemedianstrainsofRhizobium,MesorhizobiumandBradyrhizobium.
ThesefindingsphenotypicallycharacterizeSEMIA3007aspartofthegenusMesorhizobium.
ThisstrainwasoriginallyisolatedinMexico(B-11AMex)andclassifiedtaxonomic-allyasRhizobiumleguminosarumbv.
viceaeSEMIA3007byacombinationofphenotypicmethods,biochemicaltestsandpartialsequencingofthe16SrRNAgene.
GenomeassemblyofSEMIA3007anditsfeaturesThesequencingresultshowsthatstrainSEMIA3007hasthefollowingcharacteristics:onecontigof6,990,002bp,G+Ccontent63%,6,814codingsequences(CDS)andatotalof55RNAs.
IntheSEMIA3007genome,therearetwoclustersencodingnitritereductase(nirVandnirK)andfourclustersrelatedtodenitrificationprocessesthatreducenitratetonitrogengas.
Itispostu-latedthatafterhostinfectionthisclusterisresponsibleforallowingBrucellasuistosurvivelowoxygenconcen-trationsbecausethecellscanusenitrogenoxidesasfinalelectronacceptors[24,25].
ThepresenceofthispathwayenablesSEMIA3007tousethismechanismofintracellu-larsurvivalduringhostinfection.
WealsofoundthefollowingothergeneswerepresentinthegenomeofSEMIA3007:nifA,nifS,nifU,IscA-like,Fig.
1Bacterialgrowthcurve.
BradyrhizobiumelkaniiLMG6134,Rhizobiumleguminosarumbv.
viceaeLMG14904,MesorhizobiumhuakuiiLMG14107andMesorhizobiumsp.
SEMIA3007strainsKishietal.
BMCMicrobiology(2016)16:260Page2of8nifB,frdN,nifX,nifX2,nifE,nifN,nifQ,nifW,nifH,nifD,nifK,nifZandnifT.
Theseresultssuggestmechanismsfordenitrificationprocesses.
Thenifgenesfunctioninthetransformationofammonianitrogen,nitrate,andnitriteammonificationandcodeforproteinssuchasnitratereductase(EC1.
7.
99.
4),nitritereductase[NAD(P)H](EC1.
7.
1.
4),ferredoxin-nitritereductase(EC1.
7.
7.
1)andnitritereductase(cytochrome;ammonia-forming)(EC1.
7.
2.
2).
TheSEMIA3007genomealsocontainsasubsystemforassimilationofammoniaandthebacteriacanuseammoniaassimilatedformetabolismofaminoacids(glutamate).
Thesystemusesglutamateammonialigase(EC6.
3.
1.
2),glutamatesynthase(NADPH)(EC1.
4.
1.
13),glutamatesynthase(NADH)(EC1.
4.
1.
14)andglutamatesynthase(ferredoxin)(EC1.
4.
7.
1).
SystemsecretionsoftypeI,typeII,typeIVandtypeVIwereidentified.
Thesesystemcomponentsarecommonamongrhizobia.
ThetypeIVsecretionsystemsareidentifiedinmicroorganismsassociatedwithplantsandareusuallycomposedofVirproteins[26].
TheoperonofthetypeIVSEMIA3007systemfeatures12genesencodingthefollowingproteins:VirB1-VirB4,VirB6,VirB8-VirB9,VirB11,VirD4andVirG.
ThevirBregionisresponsibleforcodingkeyvirulencefactorsinthesymbiosisspeciesMesorhizobium[26,27].
Thisoperonmayassistininducingacidificationofthephagosomeinthecellsafterphagocytosis.
Theacidifica-tionmayleadtothesegregationofunknowneffectormoleculesandcreatechangesinthehostcellendosomethatgenerateanewintracellularcompartmentinwhichtheattackercanreplicate[28].
ThesecretionsystemsoftypesIII,IVandVIandthenodulationfactorsareconsideredresponsibleforleadhostspecificityinMesorhizobiumhuakuii[4].
GenomecomparisonsofSEMIA3007andRhizobiumOuranalysisofthesimilaritybetweengenomescanbeusedtodifferentiatemicroorganisms.
WeusedthegenomesofSEMIA3007,Rhizobiumleguminosarumbv.
viceae(gi115254414),Rhizobiumleguminosarumbv.
trifolii(gi240861949),Mesorhizobiumerdmanii(gi548692182),Mesorhizobiumciceri(gi317165637),Mesorhizobiumhuakuii(gi657121522),Mesorhizobiumloti(gi47118328)andMesorhizobiumopportunistum(gi336024847)toconstructaprogressivealignmentusingtheprogramMauve.
Wefoundtherewasahighdegreeofsimilarity(blocksyntenyanddirection)betweenSEMIA3007andtheMesorhizobiumgroupandalimitednumberofblockscollinearbetweenRhizobium(Fig.
2).
ANI[23]isonemethodthathasreplacedtheDDH[29],anditisthebestinsilicoparameterrepresent-ingDDHthathasbeenexperimentallydemonstrated[22,29].
OurgenomecomparisonsfortaxonomicpurposeswerebasedonBLASTcalculations[30].
AnANIvalueof95%±0.
5%identitycorrespondsto70%DDH[23],whichisavalueoftenrecommendedtodelimitspecieswhenusedinconjunctionwithothercriteria,suchasphenotypictraits[31].
RichterandRossello-Mora[17]describeasoftwaretool(JSpecies)designedtoeasilyallowthecalculationofANIbasedontheBLASTalgorithm[30]andtheMUMmerultra-rapidaligningtool[32].
Wealsocalculatedthetetranucleotidefrequencies,whicharealignment-freeparametersthathavebeensuccessfullyappliedtophylogeneticallysortmetagenomeinserts[33].
Therefore,the95–96%ANIthresholdcanbereadilyusedasanobjectiveboundaryforspeciescircumscriptionifitisreinforcedbyhighTETRAcorrelationvalues[17].
OurresultsdemonstratethatSEMIA3007ismoregeneticallysimilartoMesorhizobiumhuakuiithanRhizobium(Table1).
Phylogeneticanalysisusing16SrRNATheresultsofsequencingthe16SrRNAgeneSEMIA3007weresubjectedtoamembershipanalysistaxonomyinRDPIIbank.
Weutilizedtheclassifiertoolwithathresholdof95%.
Theresultshowedtheidentitywas100%Mesorhizobium.
Additionally,therewas100%identitywiththe16SRibosomalRNAdatabaseusingtheBlastprogram(June2006).
AphylogeneticanalysiswasperformedusingdataavailableontheNCBIdatabasetoassesswhetherSEMIA3007shouldbeidentifiedandcatalogedasRhizobiumleguminosarumbv.
viceaewithinRhizobiumorbereclassifiedaspartoftheMesorhizobiumgroup(Additionalfile1:TableS1).
TheresultsofthephylogeneticanalysisclearlyshowSEMIA3007isamemberofMesorhizobiumandisseparatefromtheRhizobiumgroup,whichsuggestsareclassificationofSEMIA3007iswarranted(Fig.
3).
ComparisonofgeneorthologsPreviousstudieshavecomparedgenestodifferentiateorganisms.
WeusedOrthoMCLclusteringtoidentify"coregenes",whicharethenumberofuniqueandsharedorthologsofSEMIA3007andMesorhizobium(Fig.
4).
Atotalof32,604proteinsfromSEMIA3007(6,814proteins),M.
huakuii(5,838proteins),M.
erdma-nii(6,491proteins),M.
loti(7,043proteins)andM.
opportunistum(6,418proteins)wereevaluated.
Weusedaninflationindexof1.
5tocompletegenesandidentified3,075orthologgroupswithinthefivegenomes.
TheclustersoforthologsinFig.
4showthereare3,075orthologgroupsinSEMIA3007representing69.
5%ofthetotalCDSinthegenome.
However,SEMIA3007andM.
huakuiishowed3,951(79.
1%)commonorthologgroups.
WefoundthatSEMIA3007Kishietal.
BMCMicrobiology(2016)16:260Page3of8andM.
erdmaniishared4,392(87.
9%)orthologs.
Therewere4,197(84%)orthologsincommonbetweenSEMIA3007andM.
loti.
Therewerealso3,984(79%)orthologssharedbetweenSEMIA3007andM.
opportu-nistum.
Therefore,isolatedSEMIA3007showsalargenumberofMesorhizobiumgeneorthologs.
Thesefind-ingssuggestthatSEMIA3007isaMesorhizobiumstrain.
Therefore,theresultsforgrowthcurveofSEMIA3007,comparativeanalysisofthegenome,ANI,geneorthologsandphylogeneticanalysisusing16SrRNAshowthatSEMIA3007isnotRhizobiumlegumi-nosarumbv.
viceaesuggestingitsreclassificationforMesorhizobiumgroup[10–13].
ConclusionsNGStechnologieshaveproventheirutilityingenomicandmetagenomicsareassincetheirearliestapplicationappearedin2006.
Identifyingeachindividualsequenceisimportantinmicrobialcommunityanalysisbecausethetaxonomicinformationprovidesaccesstobasictraitinformationsuchasphysiology,epidemiologyandevolu-tionaryhistory.
Thetaxonomicinformationalsopermitsindirectinferenceoftheirecologicalrolesinagivenenvironment[19].
Whole-genomesequencinghasproventobevaluableandcriticalforrefiningthephylogeneticpositionsandcorrecttaxonomicclassificationofrhizobialstrains[10,11,34].
Inthisstudy,wesequenced,assembledandannotatedtheSEMIA3007genome.
WeusedthisgenomesequencetoexaminethephylogeneticrelationshipbetweenMesorhizobiumandRhizobiumgenus.
SEMIA3007wasclassifiedbyphenotypicFig.
2GenomescomparisonbetweenMesorhizobiumandRhizobium.
aAgenomealignmentofeightgenomesusingMauverevealscollinearblocksconserved(LCB)amongallgenomes.
Eachchromosomeisshownhorizontallyandhomologousblocksineachgenomeareshownasidenticallycoloredregions.
bSimilaritybetweengenomesalignedbyMauveprogramshowingthephylogeneticrelationshipsbetweengenomesTable1ProbabilityofpairwisecomparisonGenome(bp)%GCGene%ANIb%ANIm%TetraSEMIA3007699000262.
97173---M.
huakuii636436563.
2583898.
4598.
9499.
97M.
loti703607162.
7704393.
4694.
6499.
96M.
opportunistum688444462.
9641887.
3189.
1799.
85M.
ciceri626448962.
5610085.
7087.
2599.
85M.
erdmanii701826562.
7649188.
8990.
4099.
86R.
leg_viciae505714261.
1479770.
7283.
5495.
75R.
leguminosarum741812260.
7729370.
2782.
7996.
01Nucleotideidentityvalues(ANI)andcorrelationindexesoftheirTetra-nucleotidesignaturesbetweenSEMIA3007withMesorhizobiumandRhizobiumKishietal.
BMCMicrobiology(2016)16:260Page4of8taxonomicmethodsandbiochemicaltestsasRhizobiumleguminosarumbv.
viceae.
However,ourresultsstronglysuggestthatSEMIA3007belongstotheMesorhizobiumgenus.
TheplacementofSEMIA3007inaMesorhizobiumgenusissupportedbyouranalysisofANI,orthologgenesandphylogeneticanalysis.
WecanseeahighdegreeofsimilarityandblocksyntenyanddirectionbetweenSEMIA3007andtheMesorhizobiumgroup.
OurresultsdemonstratedtherewerealimitednumberofblockscollinearbetweenRhizobium.
Additionally,theANIbasedonapairwisegenomecomparisonofallsharedorthologproteincodinggenesis98%withMesorhizobiumhuakuii.
OurphylogeneticanalysisdemonstratedthatSEMIA3007isnotpartoftheRhizobiumgenus,andtheorthologgenesrevealedsufficientabilitytoidentifySEMIA3007asMesorhizobium.
Theconceptsoforthologyoriginatedfromthefieldofmolecularsystematics[35]andhaverecentlybeenappliedtofunctionalcharacterizationsandclassificationsonthescaleofwhole-genomecomparisons[36–38].
Incomparativegenomics,theclusteringoforthologousgenesprovidesaframeworkforintegratinginformationfrommultiplegenomesbyhighlightingthedivergenceandconservationofgenefamiliesandbiologicalprocesses.
Theidentificationoforthologousgroupsinprokaryoticgenomeshaspermittedcross-referencingofgenesfrommultiplespeciesandhasfacilitatedgenomeannotation,proteinfamilyclassification,studiesonbacterialevolutionandtheidentificationofstrains.
Theultimategoaloftaxonomyistoconstructaclassificationthatisoperativeandpredictiveforanydisciplineinmicrobiology.
TheclassificationisalsoessentiallystableforoldandnewstrainsuchasRhizobiaandthecollectivenamesofthegeneraRhizobium,Sinorhizobium,Mesorhizobium.
MethodsBacterialgrowthcurveThestrainsofBradyrhizobiumelkaniiLMG6134,Rhizobiumleguminosarumbv.
viceaeLMG14904,MesorhizobiumhuakuiiLMG14107andMesorhizobiumsp.
SEMIA3007wereculturedfor96hwithshakingFig.
3PhylogenetictreeshowingthetaxonomicpositionofSEMIA3007strainbetweengroupsofMesorhizobiumandRhizobium.
Geneticdifferencesbetweenbacteriaof0.
4%.
ThenumbersinthebranchesshowtheprobabilitycalculatedbyMrBayeswiththecolorsKishietal.
BMCMicrobiology(2016)16:260Page5of8(150rpm)at30°CinTYmedium[39]intriplicate.
Toobtainthebacterialgrowthcurve,theODreadingwascollectedevery8h.
BacterialstrainandDNApreparationSEMIA3007wasculturedfor48hat28°Cwith145rpmshakinginTYmedium[39].
TheSEMIA3007cellswereharvestedbycentrifugation,andthetotalDNAwaspreparedusingaWizardGenomicDNAPurificationKit(Promega).
SequencingandannotationofthegenomeThedenovosequencingoftheSEMIA3007genomeusedacombinedstrategyinvolvingIllumina–HiscanSQ.
ThelibrarieswereconstructedusingaTruSeqDNASamplePrepkitandNexteraMatePairSamplePreparationkit(Illumina).
TheclusterformationoflibrarytemplateswasperformedwiththeTruSeqPEClusterkitv3(Illumina)andtheIlluminacBotworkstationusingconditionsrecommendedbythemanufacturer.
Pairedend100basepair(2x100bp)sequencingbysynthesiswasperformedwithTruSeqSBSkitv3(Illumina)onanIlluminaHiscanSQusingprotocolsdefinedbythemanufacturer.
ThebasecallconversiontosequencereadswasperformedusingCASAVA1.
8.
3(Illumina).
Asaresult,paired-endandmatepairfastqfilesweretrimmedusingScythe0.
991(https://github.
com/vsbuffalo/scythe),Cutadapt1.
7.
1[40]andthequalityofdatawasfilteredbyPrinseqprogram[41]withPhred≥20.
ThesequenceassemblywasperformedusingtheSpades3.
6.
1program[42].
Thepre-dictionofORFsandannotationwereperformedusingtheRastsystem[43].
Genomecomparisonsandaveragenucleotideidentity(ANI)ForcomparingthegenomeofSEMIA3007toothersgenomeswecomputeanalignmentofthesixgenomes,weusedtheProgressiveMauvealgorithm[44].
Analign-mentofthefourMesorhizobiumandRhizobiumgenomeswasconstructedusingthedefaultmauveAlignerparame-ters.
TheresultingLCBswereinspectedusingtheMauvealignmentviewer,andtheminimumLCBweightwasadjustedtoeliminateLCBsconsistingofonlyrepetitiveelements(LCBWeight600).
Referencegenomesforcomparisonpurposeswerere-trievedfromtheGenBankdatabase(http://www.
ncbi.
nlm.
nih.
gov/genbank/).
SequenceswereuploadedintotheJSpeciessoftwarepackage(http://www.
imedea.
uib.
es/jspecies)toperformpairwisegenomecalculationsoftheaveragenucleotideidentity(ANI)[17,23]andsupporttheproposedcut-offlevelof95%asaspeciesdelineationthreshold[22].
OrthologanalysisTheorthologgroupsinmultiplegenomescanbeusefulforannotationandrevealingthepatternsofphylogeneticproteinsfromdifferentstrains.
Thegroupsalsoprovideinsightsintotheevolutionaryconservationanddiversecellularfunctionsindifferentspecies.
Fourcodingsequences(CDS)fromgenomes/draftsofMesorhizobiumloti,Mesorhizobiumhuakuii,Mesorhizo-biumerdmaniiandMesorhizobiumsp.
wereextractedfromGenBankfiles(Additionalfile1:TableS1),repre-sentingfourspecies(fivewithSEMIA3007CDS).
Thepanandcoregenomeanalysiswasconductedbydeter-miningshared(homologous)andspecies-specificprotein-codinggenesusingOrthoMCL[36]withe-valuecutoff1*1020,proteinpercentidentity≥50%andMCLinflationof1.
5.
OrthoMCLcomputesfamiliesofhomologousgenesforpanandcoregenomeanalyses.
Thefamiliesinwhichtwoormoregenomesparticipatewereusedtodeterminenumbersplotted.
OrthoMCLwasrunwithblaste-valuecut-offof1e-5andaninfla-tionparameterof1.
5.
ThetablewithorthologswasusedtoplotVenndiagrams(http://bioinformatics.
psb.
ugent.
be/webtools/Venn/)[36].
16SrRNAgenesequencingTheamplificationofthe16SrRNAgeneoftheSEMIA3007wasperformedwithFD1andRD1primers[45].
ThePCRreactionmixtureconsistedof30ngofDNA,7.
5pmolofeachprimer,0.
2mMofdNTPs,1.
5mMofMgCl2,Buffer1Xand2.
5UTaqDNAFig.
4VenndiagramshowingcoregenomeanalysesofMesorhizobiumstrains.
Thenumberofprotein-codinggeneorthologsharingamongfiveMesorhizobium.
SEMIA3007;M.
huakuii(CP006581.
1);M.
loti(NC_002678.
2);M.
opportunistum(NC_015675.
1);M.
erdmanii(NZ_AXAE01000048.
1)Kishietal.
BMCMicrobiology(2016)16:260Page6of8polymerase(LudwigBiotec).
AthermocyclermodelPTC-100ProgrammableThermalController(MJResearch,Inc.
)wasusedwithathermalprofileof96°Cfor2min,40cyclesof96°Cfor30s,53°Cfor1minand60°Cfor4min.
AfterthePCRreaction,theproductswerepurifiedwithaWizardSVGelandPCRClean-UpSystem(Pro-mega).
Theampliconwassequencedwith1μlofBigDyeTerminatorv3.
1,buffer0.
75X(Tris-HCl200mM,pH9.
0andMgCl25mM),10pmolesofprimerFD1,50ngofDNAandsterileMilli-Qdistilledwater(10μLq.
s.
p).
SequencingwasperformedonSequen-cerABIPRISM3130xlDNAAnalyzer(AppliedBiosystems)followingthemanufacturer'sinstructions.
Downloadingthesequences16SrRNAinGenBankTheNationalCenterforBiotechnologyInformation(NCBI)wasusedtosearchthegenomeforspeciesMesorhizobium(March15,2016).
Allcompletegenese-quencesfor16SrRNA(16SribosomalRNA)weredown-loadedfromGenBank(Additionalfile1:TableS1)[46].
Phylogeneticanalysisof16SrRNAgeneThe16SrRNAgenesetwerealignedusingtheMAFFTv7.
215program[47].
ThesearchforthebestnucleotidesubstitutionmatrixwasperformedwiththePhangornpackage[48]inR[49]andthefeaturemodelTest.
TheconstructionofaphylogenetictreewasperformedwiththeMrbayesv3.
2.
2program[50]usingthematrixreplacementGeneralTimeReversible(GTR)withgammavariation(G)andinvariablesites(I)with10.
000.
000generations.
ThebestevolutionarymodelwaschosenbasedonAkaikeinformationcriterionwithcorrection(AICc).
NucleotidesequenceaccessionnumberThedatasetsresultsofthisarticleareavailableintheNCBIBioProjectSRR3703040.
AdditionalfileAdditionalfile1:TableS1.
Completegenomesanddrafts.
(XLSX18kb)Abbreviations16SrRNA:16SribosomalRNAgene;AICc:Akaikeinformationcriterionwithcorrection;ANI:Averagenucleotideidentityvalues;CDS:Codingsequences;DDH:DNA-DNAhybridization;DNA:Deoxyribonucleicacid;GTR:Generaltimereversible;LCB:Collinearblocksconserved;LCB:Locallycollinearblocks;NAD(P)H:Nitritereductase;NADH:Glutamatesynthase;NADPH:Glutamatesynthase;NCBI:NationalCenterforBiotechnologyInformation;NGS:Nextgenerationsequencing;OD:Opticaldensity;ORF:Openreadframe;PCR:Polymerasechainreaction;TY:Triptone,yeastmediumAcknowledgementsWegratefullyacknowledgeUniversidadeEstadualPaulista–UNESP,theProgramadePósGraduaoemMicrobiologiaAgropecuária,UNESP,Jaboticabal,SoPauloState,Brazil.
WealsothankFAPESP(grants:2009/539842;2014/14234-6)andCNPq.
FundingThisworkwassupportedbyFAPESP(grants:2009/539842;2014/14234-6)andCNPq.
AvailabilityofdataandmaterialsThedatasetsresultsofthisarticleareavailableintheNCBIBioProjectSRR3703040.
http://www.
ncbi.
nlm.
nih.
gov/sra/term=SRR3703040Thedatasetsresultsfromphylogenetictreeofthisarticleareavaiablein:http://purl.
org/phylo/treebase/phylows/study/TB2:S20064YoucancitethisURLinyourmanuscript.
Itwillbecomethepermanentandresolvableresourcelocatorafteryoursubmissionhasbeenapprovedandthedataaremadepublic.
http://purl.
org/phylo/treebase/phylows/study/TB2:S20064x-access-code=c3247bbe7382039c0d043d51a87de8e8&format=htmlYoucancopyandsendthisURLtoyoujournaleditortoprovidereviewerswithlimited,read-onlyaccesstoyourdata,evenifyoursubmissionhasnotyetbeenapprovedandthedataarenotyetpublic.
Authors'contributionsConceivedanddesignedtheexperiments:LTKCCFEGML.
Performedtheexperiments:LTKCCFJCCEML.
Analyzedthedata:LTKWPO.
Contributedreagents/materials/analysistools:EGML.
Wrotethepaper:CCFLTK.
Allauthorsreadandapprovedthefinalmanuscript.
CompetinginterestsTheauthorsdeclarethattheyhavenocompetinginterests.
ConsentforpublicationAllauthorsareawareofthepublication.
EthicsapprovalandconsenttoparticipateAllauthorsareawareofthepublication.
Received:29July2016Accepted:28October2016References1.
KanekoT,NakamuraY,SatoS,AsamizuE,KatoT,SasamotoS,etal.
Completegenomestructureofthenitrogen-fixingsymbioticbacteriumMesorhizobiumloti.
DNARes.
IntJRapidPublRepGenesGenomes.
2000;7:331–8.
2.
VelázquezE,PeixA,Zurdo-PiiroJL,PalomoJL,MateosPF,RivasR,etal.
Thecoexistenceofsymbiosisandpathogenicity-determininggenesinRhizobiumrhizogenesstrainsenablesthemtoinducenodulesandtumorsorhairyrootsinplants.
MolPlantMicrobeInteract.
2005;18:1325–32.
3.
GrahamPH,SadowskyMJ,KeyserHH,BarnetYM,BradleyRS,CooperJE,etal.
Proposedminimalstandardsforthedescriptionofnewgeneraandspeciesofroot-andstem-nodulatingbacteria.
IntJSystBacteriol.
1991;41:582–7.
4.
WangS,HaoB,LiJ,GuH,PengJ,XieF,etal.
Whole-genomesequencingofMesorhizobiumhuakuii7653Rprovidesmolecularinsightsintohostspecificityandsymbiosisislanddynamics.
BMCGenomics.
2014;15:440.
5.
DegefuT,Wolde-meskelE,LiuB,CleenwerckI,WillemsA,FrostegardA.
Mesorhizobiumshonensesp.
nov.
,Mesorhizobiumhawassensesp.
nov.
andMesorhizobiumabyssinicaesp.
nov.
,isolatedfromrootnodulesofdifferentagroforestrylegumetrees.
IntJSystEvolMicrobiol.
2013;63:1746–53.
6.
JarvisBDW,VanBerkumP,ChenWX,NourSM,FernandezMP,Cleyet-MarelJC,etal.
Transferofrhizobiumloti,rhizobiumhuakuii,rhizobiumciceri,rhizobiummediterraneum,andrhizobiumtianshanensetomesorhizobiumgen.
nov.
IntJSystBacteriol.
1997;47:895–8.
7.
MousaviSA,WillemsA,NesmeX,deLajudieP,LindstrmK.
Revisedphylogenyofrhizobiaceae:proposalofthedelineationofpararhizobiumgen.
nov.
,and13newspeciescombinations.
SystApplMicrobiol.
2015;38:84–90.
8.
MousaviSA,stermanJ,WahlbergN,NesmeX,LavireC,VialL,etal.
PhylogenyoftheRhizobium–Allorhizobium–AgrobacteriumcladesupportsthedelineationofNeorhizobiumgen.
nov.
SystApplMicrobiol.
2014;37:208–15.
9.
Martínez-HidalgoP,Martínez-MolinaE,MateosPF,VelázquezE,Peix,Flores-FélixJD,etal.
RevisionofthetaxonomicstatusoftypestrainsofKishietal.
BMCMicrobiology(2016)16:260Page7of8MesorhizobiumlotiandreclassificationofstrainUSDA3471TasthetypestrainofMesorhizobiumerdmaniisp.
nov.
andATCC33669TasthetypestrainofMesorhizobiumjarvisiisp.
nov.
Int.
J.
Syst.
Evol.
Microbiol.
2015;65:1703–8.
10.
Ormeo-OrrilloE,Servín-GarcidueasLE,RogelMA,GonzálezV,PeraltaH,MoraJ,etal.
TaxonomyofrhizobiaandagrobacteriafromtheRhizobiaceaefamilyinlightofgenomics.
SystApplMicrobiol.
2015;38:287–91.
11.
GuptaA,SharmaVK.
Usingthetaxon-specificgenesforthetaxonomicclassificationofbacterialgenomes.
BMCGenomics[Internet].
2015[cited2016Oct4];16.
Availablefrom:http://www.
biomedcentral.
com/1471-2164/16/396.
Accessed5Oct2016.
12.
TengJLL,TangY,HuangY,GuoF-B,WeiW,ChenJHK,etal.
PhylogenomicAnalysesandReclassificationofSpecieswithintheGenusTsukamurella:InsightstoSpeciesDefinitioninthePost-genomicEra.
Front.
Microbiol.
[Internet].
2016[cited2016Oct4];7.
Availablefrom:http://journal.
frontiersin.
org/Article/10.
3389/fmicb.
2016.
01137/abstract.
Accessed5Oct2016.
13.
McIlroySJ,LapidusA,ThomsenTR,HanJ,HaynesM,LobosE,etal.
HighqualitydraftgenomesequenceofMeganemaperideroedesstr.
Gr1TandaproposalforitsreclassificationtothefamilyMeganemaceaefam.
nov.
Stand.
GenomicSci.
[Internet].
2015[cited2016Oct4];10.
Availablefrom:http://www.
standardsingenomics.
com/content/10/1/23.
Accessed5Oct2016.
14.
VandammeP,PotB,GillisM,deVosP,KerstersK,SwingsJ.
Polyphasictaxonomy,aconsensusapproachtobacterialsystematics.
MicrobiolRev.
1996;60:407–38.
15.
KimB-Y,WeonH-Y,CousinS,YooS-H,KwonS-W,GoS-J,etal.
Flavobacteriumdaejeonensesp.
nov.
andFlavobacteriumsuncheonensesp.
nov.
,isolatedfromgreenhousesoilsinKorea.
IntJSystEvolMicrobiol.
2006;56:1645–9.
16.
MennaP,HungriaM,BarcellosFG,BangelEV,HessPN,Martínez-RomeroE.
Molecularphylogenybasedonthe16SrRNAgeneofeliterhizobialstrainsusedinBraziliancommercialinoculants.
SystApplMicrobiol.
2006;29:315–32.
17.
RichterM,Rossello-MoraR.
Shiftingthegenomicgoldstandardfortheprokaryoticspeciesdefinition.
ProcNatlAcadSci.
2009;106:19126–31.
18.
CoenyeT,GeversD,VandePeerY,VandammeP,SwingsJ.
Towardsaprokaryoticgenomictaxonomy.
FEMSMicrobiolRev.
2005;29:147–67.
19.
KimM,LeeK-H,YoonS-W,KimB-S,ChunJ,YiH.
Analyticaltoolsanddatabasesformetagenomicsinthenext-generationsequencingera.
GenomicsInform.
2013;11:102.
20.
AchtmanM,WagnerM.
Microbialdiversityandthegeneticnatureofmicrobialspecies.
Nat.
Rev.
Microbiol.
[Internet].
2008[cited2016Jun22];Availablefrom:http://www.
nature.
com/nrmicro/journal/v6/n6/abs/nrmicro1872.
html.
Accessed5Oct2016.
21.
KonstantinidisKT,TiedjeJM.
Genomicinsightsthatadvancethespeciesdefinitionforprokaryotes.
ProcNatlAcadSci.
2005;102:2567–72.
22.
KonstantinidisKT,TiedjeJM.
Trendsbetweengenecontentandgenomesizeinprokaryoticspecieswithlargergenomes.
ProcNatlAcadSciUSA.
2004;101:3160–5.
23.
GorisJ,KlappenbachJA,VandammeP,CoenyeT,KonstantinidisKT,TiedjeJM.
DNA–DNAhybridizationvaluesandtheirrelationshiptowhole-genomesequencesimilarities.
IntJSystEvolMicrobiol.
2007;57:81–91.
24.
KohlerS,FoulongneV,Ouahrani-BettacheS,BourgG,TeyssierJ,RamuzM,etal.
Nonlinearpartialdifferentialequationsandapplications:theanalysisoftheintramacrophagicvirulomeofBrucellasuisdecipherstheenvironmentencounteredbythepathogeninsidethemacrophagehostcell.
ProcNatlAcadSci.
2002;99:15711–6.
25.
HaineV,DozotM,DornandJ,LetessonJ-J,DeBolleX.
NnrAisrequiredforfullvirulenceandregulatesseveralbrucellamelitensisdenitrificationgenes.
JBacteriol.
2006;188:1615–9.
26.
HubberAM,SullivanJT,RonsonCW.
Symbiosis-inducedcascaderegulationoftheMesorhizobiumlotiR7AVirB/D4typeIVsecretionsystem.
MolPlantMicrobeInteract.
2007;20:255–61.
27.
SotoMJ,SanjuanJ,OlivaresJ.
Rhizobiaandplant-pathogenicbacteria:commoninfectionweapons.
Microbiology.
2006;152:3167–74.
28.
BoschiroliML,Ouahrani-BettacheS,FoulongneV,Michaux-CharachonS,BourgG,Allardet-ServentA,etal.
TypeIVsecretionandBrucellavirulence.
VetMicrobiol.
2002;90:341–8.
29.
GarrityGM,TrüperHG,WhitmanWB,GrimontPAD,NesmeX,FrederiksenW,etal.
Reportoftheadhoccommitteeforthere-evaluationofthespeciesdefinitioninbacteriology.
IntJSystEvolMicrobiol.
2002;52:1043–7.
30.
AltschulSF,GishW,MillerW,MyersEW,LipmanDJ.
Basiclocalalignmentsearchtool.
JMolBiol.
1990;215:403–10.
31.
Rosselló-MoraR,AmannR.
Thespeciesconceptforprokaryotes.
FEMSMicrobiolRev.
2001;25:39–67.
32.
DelcherAL,KasifS,FleischmannRD,PetersonJ,WhiteO,SalzbergSL.
Alignmentofwholegenomes.
NucleicAcidsRes.
1999;27:2369–76.
33.
TeelingH,MeyerdierksA,BauerM,AmannR,GlocknerFO.
Applicationoftetranucleotidefrequenciesfortheassignmentofgenomicfragments.
EnvironMicrobiol.
2004;6:938–47.
34.
SchuldesJ,RodriguezOrbegosoM,SchmeisserC,KrishnanHB,DanielR,StreitWR.
Completegenomesequenceofthebroad-host-rangestrainsinorhizobiumfrediiUSDA257.
JBacteriol.
2012;194:4483.
35.
FitchWM.
Distinguishinghomologousfromanalogousproteins.
SystZool.
1970;19:99.
36.
LiL.
OrthoMCL:identificationoforthologgroupsforeukaryoticgenomes.
GenomeRes.
2003;13:2178–89.
37.
TatusovRL,KooninEV,LipmanDJ.
Agenomicperspectiveonproteinfamilies.
Science.
1997;278:631–7.
38.
ChervitzSA,AravindL,SherlockG,BallCA,KooninEV,DwightSS,etal.
Comparisonofthecompleteproteinsetsofwormandyeast:orthologyanddivergence.
Science.
1998;282:2022–8.
39.
BeringerJE.
RfactortransferinRhizobiumleguminosarum.
JGenMicrobiol.
1974;84:188–98.
40.
MartinM.
Cutadaptremovesadaptersequencesfromhigh-throughputsequencingreads.
EMBnetJ.
2011;17:10.
41.
SchmiederR,EdwardsR.
Qualitycontrolandpreprocessingofmetagenomicdatasets.
Bioinformatics.
2011;27:863–4.
42.
BankevichA,NurkS,AntipovD,GurevichAA,DvorkinM,KulikovAS,etal.
SPAdes:anewgenomeassemblyalgorithmanditsapplicationstosingle-cellsequencing.
JComputBiol.
2012;19:455–77.
43.
AzizRK,BartelsD,BestAA,DeJonghM,DiszT,EdwardsRA,etal.
TheRASTserver:rapidannotationsusingsubsystemstechnology.
BMCGenomics.
2008;9:75.
44.
DarlingACE.
Mauve:multiplealignmentofconservedgenomicsequencewithrearrangements.
GenomeRes.
2004;14:1394–403.
45.
WeisburgWG,BarnsSM,PelletierDA,LaneDJ.
16SribosomalDNAamplificationforphylogeneticstudy.
JBacteriol.
1991;173:697–703.
46.
LaranjoM,AlexandreA,OliveiraS.
Legumegrowth-promotingrhizobia:anoverviewontheMesorhizobiumgenus.
MicrobiolRes.
2014;169:2–17.
47.
KatohK.
MAFFT:anovelmethodforrapidmultiplesequencealignmentbasedonfastFouriertransform.
NucleicAcidsRes.
2002;30:3059–66.
48.
SchliepKP.
phangorn:phylogeneticanalysisinR.
Bioinformatics.
2011;27:592–3.
49.
IhakaR,GentlemanR.
R:Alanguagefordataanalysisandgraphics.
JComputGraphStat.
1995;5:299–314.
50.
RonquistF,TeslenkoM,vanderMarkP,AyresDL,DarlingA,HohnaS,etal.
MrBayes3.
2:efficientbayesianphylogeneticinferenceandmodelchoiceacrossalargemodelspace.
SystBiol.
2012;61:539–42.
Weacceptpre-submissioninquiriesOurselectortoolhelpsyoutondthemostrelevantjournalWeprovideroundtheclockcustomersupportConvenientonlinesubmissionThoroughpeerreviewInclusioninPubMedandallmajorindexingservicesMaximumvisibilityforyourresearchSubmityourmanuscriptatwww.
biomedcentral.
com/submitSubmityournextmanuscripttoBioMedCentralandwewillhelpyouateverystep:Kishietal.
BMCMicrobiology(2016)16:260Page8of8
viceaebybioinformatictoolsLucianoTakeshiKishi,CamilaCesárioFernandes,WellingtonPineOmori,JooCarlosCampanharoandElianaGertrudesdeMacedoLemos*AbstractBackground:EvidencebasedongenomicsequencesisextremelyimportanttoconfirmthephylogeneticrelationshipswithintheRhizobiumgroup.
SEMIA3007wasanalyzedwithintheMesorhizobiumgroupstodefinetheunderlyingcausesoftaxonomicidentification.
Wepreviouslyusedbiochemicaltestsandphenotypictaxonomicmethodstoidentifybacteria,whichcanleadtoerroneousclassification.
AnimprovedunderstandingofbacterialstrainssuchastheMesorhizobiumgenuswouldincreaseourknowledgeofclassificationandevolutionofthesespecies.
Results:Inthisstudy,wesequencedthecompletegenomeofSEMIA3007andcompareditwithfiveotherMesorhizobiumandtwoRhizobiumgenomes.
ThegenomesofisolatedSEMIA3007showedseveralorthologswithM.
huakuii,M.
erdmaniiandM.
loti.
WeidentifiedSEMIA3007asaMesorhizobiumbycomparingthe16SrRNAgeneandthecompletegenome.
Conclusion:Ourortholog,16SrRNAgeneandaveragenucleotideidentityvalues(ANI)analysisalldemonstrateSEMIA3007isnotRhizobiumleguminosarumbv.
viceae.
TheresultsofthephylogeneticanalysisclearlyshowSEMIA3007ispartoftheMesorhizobiumgroupandsuggestareclassificationiswarranted.
Keywords:Genomesequencing,Coregenome,Comparativeanalysis,Orthologgenes,PhylogeneticanalysisBackgroundRhizobiaisthecollectivenameofthegeneraRhizobium,SinorhizobiumandMesorhizobium,whicharesoilandrhizospherebacteriaofagronomicimportancebecausetheyformnitrogen-fixingsymbioseswithleguminousplants[1,2].
Thus,rhizobiaareconsideredbio-fertilizersandhavebeenusedasinoculantsforover120years.
Rhizobialgeneticdiversityandtheplant-bacteriamo-lecularinteractionshavebeenwell-studied[3].
ThegrowthrateofMesorhizobiumisintermediatebetweenthegeneraRhizobiumandBradyrhizobiumandisoneofthelargestgenera.
Additionally,theMesorhizobiumgeneraconsistsof24speciesfoundinAsia,Europe,theMediterraneanregionandAfrica[4,5].
Jarvinsetal.
[6]werethefirsttorequestthecreationoftheMesorhizobiumgenusandreclassifiedseveralgeneraidentifiedasRhizobiumintoMesorhizobium.
Thecorrectphylogeneticidentificationofaspeciesrequiresanaccuratetechnicalcharacterization[7,8].
ThetaxonomyofMesorhizobiumrequiresthereclassi-ficationofspeciesbecausethereisaneedforstudiestoavoidclassificationproblems.
Taxonomicinformationprovidesaccesstobasictraitinformationsuchasphysi-ology,epidemiologyandevolutionaryhistory[9].
Thecorrecttaxonomicassignmentofbacterialgenomesisaprimaryandchallengingtask[10–13].
*Correspondence:egerle@fcav.
unesp.
brEqualcontributorsDepartamentodeTecnologia,LaboratóriodeBioquímicadeMicrorganismosePlanta–LBMP,UNESP-UniversidadeEstadualPaulista,FaculdadedeCiênciasAgráriaseVeterinárias,ViadeAcessoProf.
PauloDonatoCastellanes/n,14884-900Jaboticabal,SP,BrazilTheAuthor(s).
2016OpenAccessThisarticleisdistributedunderthetermsoftheCreativeCommonsAttribution4.
0InternationalLicense(http://creativecommons.
org/licenses/by/4.
0/),whichpermitsunrestricteduse,distribution,andreproductioninanymedium,providedyougiveappropriatecredittotheoriginalauthor(s)andthesource,providealinktotheCreativeCommonslicense,andindicateifchangesweremade.
TheCreativeCommonsPublicDomainDedicationwaiver(http://creativecommons.
org/publicdomain/zero/1.
0/)appliestothedatamadeavailableinthisarticle,unlessotherwisestated.
Kishietal.
BMCMicrobiology(2016)16:260DOI10.
1186/s12866-016-0882-5Thepartial16SribosomalRNAgene(16SrRNA)isamolecularmarkerwidelyusedinthetaxonomyofbacteria.
However,thisgenehasnoconsensussequencetocorrectlyclassifymicroorganismsatthespecieslevel[14–16].
Thus,DNA-DNAhybridization(DDH)hasbeenusedasthegoldstandardfordefiningprokaryoticspeciesatthegenomiclevel.
DDHistheonlytaxonomicmethodthatoffersanumericalandrelativelystablespecies.
Therefore,DDHinfluenceshowthecurrentclassificationsystemhasbeenconstructed[17].
DDHisanexpensiveandlaboriousmethodthatisavailableinonlyafewlaboratoriesworldwide,sinceitrequiresthehybridizationofhundredsofstrainsandoftendoesnotresolvethetaxonomicproblems.
How-ever,itisanimportantlimitingfactorforthedescriptionofnewspecies,particularlyincountrieswiththegreatestbiodiversity.
ProkaryoticspeciescontinuetobeagroupofstrainsduetoDNA-DNAre-associationvaluesgreaterthan70%[14,18].
Therecentdevelopmentofsequencingtechnologieshasenabledustocarefullyassessmicrobialcommunitiesbygeneratingmanynucleotidesequencesatlowercosts.
Nextgenerationsequencing(NGS)technologieshaverevolutionizedthefieldofmicrobialecologyandallowsresearcherstodeterminethelevelofdiversitymorecloselyusingin-depthsequencing.
TherearevariousapplicationsusingtheseNGSplatforms,whichrangefromsingle-genetargetedsequencingtowhole-genomesequencingandshotgunmetagenomesequencing[19].
Withtheavailabilityofwholegenomesequences,thegenecontentbasedapproachesappearpromisingininferringthebacterialtaxonomy.
Thecompletegenomesequencingofabacterialgenomeoftenrevealsasubstantialnumberofuniquegenespresentonlyinthatgenomewhichcanbeusedforitstaxonomicclassification[11,12].
TherecentimprovedaccesstovariousnewgenesequencesandthedefinitionofprokaryotespecieshasledtodoubtsregardingthesuitabilityoftheDNA-DNAhybridizationmethod[20].
Thenewproposalsincludetheanalysisofseveralgenesortheentiregenome.
Oneproposedanalysismethodistoanalyzecommongenesbetweentwostrainsanddeterminetheaveragenucleo-tideidentityvalues(ANI).
AnANIvalueexceeding94%correspondsto70%traditionalDNA-DNAhybridization[21,22].
Thisanalysismethodalsoconsidersgeneswitheco-logicalfunctions.
OtherANIvaluessuggestedreplacing70%hybridizationwith95%ANIand69%conservedDNA.
Intheproteincodingportionofthegenome,thesevalueswouldsuggest85%conservedgenes[23].
Themostrecentproposalsrecommend>95–96%ANItodelineatespeciesandwouldreplacethetraditional70%cutoffthresholdusedforDDHsequences[17].
TheaimofthisstudywastoevaluateSEMIA3007isolatedinMexicoasB-11AMexandisclassifiedbyphenotypictaxonomicmethodssuchasRhizobiumlegu-minosarumbv.
viceaebydifferentgroupsofresearchers.
Weusedacombinationofcomplete16SrRNAsequen-cingandcompletegenomeanalysistoreclassifyB-11AasMesorhizobiumsp.
ResultsanddiscussionBacterialgrowthcurveThebacterialgrowthcurveofSEMIA3007isshowninFig.
1.
SEMIA3007grewsimilartothemedianstrainsofRhizobium,MesorhizobiumandBradyrhizobium.
ThesefindingsphenotypicallycharacterizeSEMIA3007aspartofthegenusMesorhizobium.
ThisstrainwasoriginallyisolatedinMexico(B-11AMex)andclassifiedtaxonomic-allyasRhizobiumleguminosarumbv.
viceaeSEMIA3007byacombinationofphenotypicmethods,biochemicaltestsandpartialsequencingofthe16SrRNAgene.
GenomeassemblyofSEMIA3007anditsfeaturesThesequencingresultshowsthatstrainSEMIA3007hasthefollowingcharacteristics:onecontigof6,990,002bp,G+Ccontent63%,6,814codingsequences(CDS)andatotalof55RNAs.
IntheSEMIA3007genome,therearetwoclustersencodingnitritereductase(nirVandnirK)andfourclustersrelatedtodenitrificationprocessesthatreducenitratetonitrogengas.
Itispostu-latedthatafterhostinfectionthisclusterisresponsibleforallowingBrucellasuistosurvivelowoxygenconcen-trationsbecausethecellscanusenitrogenoxidesasfinalelectronacceptors[24,25].
ThepresenceofthispathwayenablesSEMIA3007tousethismechanismofintracellu-larsurvivalduringhostinfection.
WealsofoundthefollowingothergeneswerepresentinthegenomeofSEMIA3007:nifA,nifS,nifU,IscA-like,Fig.
1Bacterialgrowthcurve.
BradyrhizobiumelkaniiLMG6134,Rhizobiumleguminosarumbv.
viceaeLMG14904,MesorhizobiumhuakuiiLMG14107andMesorhizobiumsp.
SEMIA3007strainsKishietal.
BMCMicrobiology(2016)16:260Page2of8nifB,frdN,nifX,nifX2,nifE,nifN,nifQ,nifW,nifH,nifD,nifK,nifZandnifT.
Theseresultssuggestmechanismsfordenitrificationprocesses.
Thenifgenesfunctioninthetransformationofammonianitrogen,nitrate,andnitriteammonificationandcodeforproteinssuchasnitratereductase(EC1.
7.
99.
4),nitritereductase[NAD(P)H](EC1.
7.
1.
4),ferredoxin-nitritereductase(EC1.
7.
7.
1)andnitritereductase(cytochrome;ammonia-forming)(EC1.
7.
2.
2).
TheSEMIA3007genomealsocontainsasubsystemforassimilationofammoniaandthebacteriacanuseammoniaassimilatedformetabolismofaminoacids(glutamate).
Thesystemusesglutamateammonialigase(EC6.
3.
1.
2),glutamatesynthase(NADPH)(EC1.
4.
1.
13),glutamatesynthase(NADH)(EC1.
4.
1.
14)andglutamatesynthase(ferredoxin)(EC1.
4.
7.
1).
SystemsecretionsoftypeI,typeII,typeIVandtypeVIwereidentified.
Thesesystemcomponentsarecommonamongrhizobia.
ThetypeIVsecretionsystemsareidentifiedinmicroorganismsassociatedwithplantsandareusuallycomposedofVirproteins[26].
TheoperonofthetypeIVSEMIA3007systemfeatures12genesencodingthefollowingproteins:VirB1-VirB4,VirB6,VirB8-VirB9,VirB11,VirD4andVirG.
ThevirBregionisresponsibleforcodingkeyvirulencefactorsinthesymbiosisspeciesMesorhizobium[26,27].
Thisoperonmayassistininducingacidificationofthephagosomeinthecellsafterphagocytosis.
Theacidifica-tionmayleadtothesegregationofunknowneffectormoleculesandcreatechangesinthehostcellendosomethatgenerateanewintracellularcompartmentinwhichtheattackercanreplicate[28].
ThesecretionsystemsoftypesIII,IVandVIandthenodulationfactorsareconsideredresponsibleforleadhostspecificityinMesorhizobiumhuakuii[4].
GenomecomparisonsofSEMIA3007andRhizobiumOuranalysisofthesimilaritybetweengenomescanbeusedtodifferentiatemicroorganisms.
WeusedthegenomesofSEMIA3007,Rhizobiumleguminosarumbv.
viceae(gi115254414),Rhizobiumleguminosarumbv.
trifolii(gi240861949),Mesorhizobiumerdmanii(gi548692182),Mesorhizobiumciceri(gi317165637),Mesorhizobiumhuakuii(gi657121522),Mesorhizobiumloti(gi47118328)andMesorhizobiumopportunistum(gi336024847)toconstructaprogressivealignmentusingtheprogramMauve.
Wefoundtherewasahighdegreeofsimilarity(blocksyntenyanddirection)betweenSEMIA3007andtheMesorhizobiumgroupandalimitednumberofblockscollinearbetweenRhizobium(Fig.
2).
ANI[23]isonemethodthathasreplacedtheDDH[29],anditisthebestinsilicoparameterrepresent-ingDDHthathasbeenexperimentallydemonstrated[22,29].
OurgenomecomparisonsfortaxonomicpurposeswerebasedonBLASTcalculations[30].
AnANIvalueof95%±0.
5%identitycorrespondsto70%DDH[23],whichisavalueoftenrecommendedtodelimitspecieswhenusedinconjunctionwithothercriteria,suchasphenotypictraits[31].
RichterandRossello-Mora[17]describeasoftwaretool(JSpecies)designedtoeasilyallowthecalculationofANIbasedontheBLASTalgorithm[30]andtheMUMmerultra-rapidaligningtool[32].
Wealsocalculatedthetetranucleotidefrequencies,whicharealignment-freeparametersthathavebeensuccessfullyappliedtophylogeneticallysortmetagenomeinserts[33].
Therefore,the95–96%ANIthresholdcanbereadilyusedasanobjectiveboundaryforspeciescircumscriptionifitisreinforcedbyhighTETRAcorrelationvalues[17].
OurresultsdemonstratethatSEMIA3007ismoregeneticallysimilartoMesorhizobiumhuakuiithanRhizobium(Table1).
Phylogeneticanalysisusing16SrRNATheresultsofsequencingthe16SrRNAgeneSEMIA3007weresubjectedtoamembershipanalysistaxonomyinRDPIIbank.
Weutilizedtheclassifiertoolwithathresholdof95%.
Theresultshowedtheidentitywas100%Mesorhizobium.
Additionally,therewas100%identitywiththe16SRibosomalRNAdatabaseusingtheBlastprogram(June2006).
AphylogeneticanalysiswasperformedusingdataavailableontheNCBIdatabasetoassesswhetherSEMIA3007shouldbeidentifiedandcatalogedasRhizobiumleguminosarumbv.
viceaewithinRhizobiumorbereclassifiedaspartoftheMesorhizobiumgroup(Additionalfile1:TableS1).
TheresultsofthephylogeneticanalysisclearlyshowSEMIA3007isamemberofMesorhizobiumandisseparatefromtheRhizobiumgroup,whichsuggestsareclassificationofSEMIA3007iswarranted(Fig.
3).
ComparisonofgeneorthologsPreviousstudieshavecomparedgenestodifferentiateorganisms.
WeusedOrthoMCLclusteringtoidentify"coregenes",whicharethenumberofuniqueandsharedorthologsofSEMIA3007andMesorhizobium(Fig.
4).
Atotalof32,604proteinsfromSEMIA3007(6,814proteins),M.
huakuii(5,838proteins),M.
erdma-nii(6,491proteins),M.
loti(7,043proteins)andM.
opportunistum(6,418proteins)wereevaluated.
Weusedaninflationindexof1.
5tocompletegenesandidentified3,075orthologgroupswithinthefivegenomes.
TheclustersoforthologsinFig.
4showthereare3,075orthologgroupsinSEMIA3007representing69.
5%ofthetotalCDSinthegenome.
However,SEMIA3007andM.
huakuiishowed3,951(79.
1%)commonorthologgroups.
WefoundthatSEMIA3007Kishietal.
BMCMicrobiology(2016)16:260Page3of8andM.
erdmaniishared4,392(87.
9%)orthologs.
Therewere4,197(84%)orthologsincommonbetweenSEMIA3007andM.
loti.
Therewerealso3,984(79%)orthologssharedbetweenSEMIA3007andM.
opportu-nistum.
Therefore,isolatedSEMIA3007showsalargenumberofMesorhizobiumgeneorthologs.
Thesefind-ingssuggestthatSEMIA3007isaMesorhizobiumstrain.
Therefore,theresultsforgrowthcurveofSEMIA3007,comparativeanalysisofthegenome,ANI,geneorthologsandphylogeneticanalysisusing16SrRNAshowthatSEMIA3007isnotRhizobiumlegumi-nosarumbv.
viceaesuggestingitsreclassificationforMesorhizobiumgroup[10–13].
ConclusionsNGStechnologieshaveproventheirutilityingenomicandmetagenomicsareassincetheirearliestapplicationappearedin2006.
Identifyingeachindividualsequenceisimportantinmicrobialcommunityanalysisbecausethetaxonomicinformationprovidesaccesstobasictraitinformationsuchasphysiology,epidemiologyandevolu-tionaryhistory.
Thetaxonomicinformationalsopermitsindirectinferenceoftheirecologicalrolesinagivenenvironment[19].
Whole-genomesequencinghasproventobevaluableandcriticalforrefiningthephylogeneticpositionsandcorrecttaxonomicclassificationofrhizobialstrains[10,11,34].
Inthisstudy,wesequenced,assembledandannotatedtheSEMIA3007genome.
WeusedthisgenomesequencetoexaminethephylogeneticrelationshipbetweenMesorhizobiumandRhizobiumgenus.
SEMIA3007wasclassifiedbyphenotypicFig.
2GenomescomparisonbetweenMesorhizobiumandRhizobium.
aAgenomealignmentofeightgenomesusingMauverevealscollinearblocksconserved(LCB)amongallgenomes.
Eachchromosomeisshownhorizontallyandhomologousblocksineachgenomeareshownasidenticallycoloredregions.
bSimilaritybetweengenomesalignedbyMauveprogramshowingthephylogeneticrelationshipsbetweengenomesTable1ProbabilityofpairwisecomparisonGenome(bp)%GCGene%ANIb%ANIm%TetraSEMIA3007699000262.
97173---M.
huakuii636436563.
2583898.
4598.
9499.
97M.
loti703607162.
7704393.
4694.
6499.
96M.
opportunistum688444462.
9641887.
3189.
1799.
85M.
ciceri626448962.
5610085.
7087.
2599.
85M.
erdmanii701826562.
7649188.
8990.
4099.
86R.
leg_viciae505714261.
1479770.
7283.
5495.
75R.
leguminosarum741812260.
7729370.
2782.
7996.
01Nucleotideidentityvalues(ANI)andcorrelationindexesoftheirTetra-nucleotidesignaturesbetweenSEMIA3007withMesorhizobiumandRhizobiumKishietal.
BMCMicrobiology(2016)16:260Page4of8taxonomicmethodsandbiochemicaltestsasRhizobiumleguminosarumbv.
viceae.
However,ourresultsstronglysuggestthatSEMIA3007belongstotheMesorhizobiumgenus.
TheplacementofSEMIA3007inaMesorhizobiumgenusissupportedbyouranalysisofANI,orthologgenesandphylogeneticanalysis.
WecanseeahighdegreeofsimilarityandblocksyntenyanddirectionbetweenSEMIA3007andtheMesorhizobiumgroup.
OurresultsdemonstratedtherewerealimitednumberofblockscollinearbetweenRhizobium.
Additionally,theANIbasedonapairwisegenomecomparisonofallsharedorthologproteincodinggenesis98%withMesorhizobiumhuakuii.
OurphylogeneticanalysisdemonstratedthatSEMIA3007isnotpartoftheRhizobiumgenus,andtheorthologgenesrevealedsufficientabilitytoidentifySEMIA3007asMesorhizobium.
Theconceptsoforthologyoriginatedfromthefieldofmolecularsystematics[35]andhaverecentlybeenappliedtofunctionalcharacterizationsandclassificationsonthescaleofwhole-genomecomparisons[36–38].
Incomparativegenomics,theclusteringoforthologousgenesprovidesaframeworkforintegratinginformationfrommultiplegenomesbyhighlightingthedivergenceandconservationofgenefamiliesandbiologicalprocesses.
Theidentificationoforthologousgroupsinprokaryoticgenomeshaspermittedcross-referencingofgenesfrommultiplespeciesandhasfacilitatedgenomeannotation,proteinfamilyclassification,studiesonbacterialevolutionandtheidentificationofstrains.
Theultimategoaloftaxonomyistoconstructaclassificationthatisoperativeandpredictiveforanydisciplineinmicrobiology.
TheclassificationisalsoessentiallystableforoldandnewstrainsuchasRhizobiaandthecollectivenamesofthegeneraRhizobium,Sinorhizobium,Mesorhizobium.
MethodsBacterialgrowthcurveThestrainsofBradyrhizobiumelkaniiLMG6134,Rhizobiumleguminosarumbv.
viceaeLMG14904,MesorhizobiumhuakuiiLMG14107andMesorhizobiumsp.
SEMIA3007wereculturedfor96hwithshakingFig.
3PhylogenetictreeshowingthetaxonomicpositionofSEMIA3007strainbetweengroupsofMesorhizobiumandRhizobium.
Geneticdifferencesbetweenbacteriaof0.
4%.
ThenumbersinthebranchesshowtheprobabilitycalculatedbyMrBayeswiththecolorsKishietal.
BMCMicrobiology(2016)16:260Page5of8(150rpm)at30°CinTYmedium[39]intriplicate.
Toobtainthebacterialgrowthcurve,theODreadingwascollectedevery8h.
BacterialstrainandDNApreparationSEMIA3007wasculturedfor48hat28°Cwith145rpmshakinginTYmedium[39].
TheSEMIA3007cellswereharvestedbycentrifugation,andthetotalDNAwaspreparedusingaWizardGenomicDNAPurificationKit(Promega).
SequencingandannotationofthegenomeThedenovosequencingoftheSEMIA3007genomeusedacombinedstrategyinvolvingIllumina–HiscanSQ.
ThelibrarieswereconstructedusingaTruSeqDNASamplePrepkitandNexteraMatePairSamplePreparationkit(Illumina).
TheclusterformationoflibrarytemplateswasperformedwiththeTruSeqPEClusterkitv3(Illumina)andtheIlluminacBotworkstationusingconditionsrecommendedbythemanufacturer.
Pairedend100basepair(2x100bp)sequencingbysynthesiswasperformedwithTruSeqSBSkitv3(Illumina)onanIlluminaHiscanSQusingprotocolsdefinedbythemanufacturer.
ThebasecallconversiontosequencereadswasperformedusingCASAVA1.
8.
3(Illumina).
Asaresult,paired-endandmatepairfastqfilesweretrimmedusingScythe0.
991(https://github.
com/vsbuffalo/scythe),Cutadapt1.
7.
1[40]andthequalityofdatawasfilteredbyPrinseqprogram[41]withPhred≥20.
ThesequenceassemblywasperformedusingtheSpades3.
6.
1program[42].
Thepre-dictionofORFsandannotationwereperformedusingtheRastsystem[43].
Genomecomparisonsandaveragenucleotideidentity(ANI)ForcomparingthegenomeofSEMIA3007toothersgenomeswecomputeanalignmentofthesixgenomes,weusedtheProgressiveMauvealgorithm[44].
Analign-mentofthefourMesorhizobiumandRhizobiumgenomeswasconstructedusingthedefaultmauveAlignerparame-ters.
TheresultingLCBswereinspectedusingtheMauvealignmentviewer,andtheminimumLCBweightwasadjustedtoeliminateLCBsconsistingofonlyrepetitiveelements(LCBWeight600).
Referencegenomesforcomparisonpurposeswerere-trievedfromtheGenBankdatabase(http://www.
ncbi.
nlm.
nih.
gov/genbank/).
SequenceswereuploadedintotheJSpeciessoftwarepackage(http://www.
imedea.
uib.
es/jspecies)toperformpairwisegenomecalculationsoftheaveragenucleotideidentity(ANI)[17,23]andsupporttheproposedcut-offlevelof95%asaspeciesdelineationthreshold[22].
OrthologanalysisTheorthologgroupsinmultiplegenomescanbeusefulforannotationandrevealingthepatternsofphylogeneticproteinsfromdifferentstrains.
Thegroupsalsoprovideinsightsintotheevolutionaryconservationanddiversecellularfunctionsindifferentspecies.
Fourcodingsequences(CDS)fromgenomes/draftsofMesorhizobiumloti,Mesorhizobiumhuakuii,Mesorhizo-biumerdmaniiandMesorhizobiumsp.
wereextractedfromGenBankfiles(Additionalfile1:TableS1),repre-sentingfourspecies(fivewithSEMIA3007CDS).
Thepanandcoregenomeanalysiswasconductedbydeter-miningshared(homologous)andspecies-specificprotein-codinggenesusingOrthoMCL[36]withe-valuecutoff1*1020,proteinpercentidentity≥50%andMCLinflationof1.
5.
OrthoMCLcomputesfamiliesofhomologousgenesforpanandcoregenomeanalyses.
Thefamiliesinwhichtwoormoregenomesparticipatewereusedtodeterminenumbersplotted.
OrthoMCLwasrunwithblaste-valuecut-offof1e-5andaninfla-tionparameterof1.
5.
ThetablewithorthologswasusedtoplotVenndiagrams(http://bioinformatics.
psb.
ugent.
be/webtools/Venn/)[36].
16SrRNAgenesequencingTheamplificationofthe16SrRNAgeneoftheSEMIA3007wasperformedwithFD1andRD1primers[45].
ThePCRreactionmixtureconsistedof30ngofDNA,7.
5pmolofeachprimer,0.
2mMofdNTPs,1.
5mMofMgCl2,Buffer1Xand2.
5UTaqDNAFig.
4VenndiagramshowingcoregenomeanalysesofMesorhizobiumstrains.
Thenumberofprotein-codinggeneorthologsharingamongfiveMesorhizobium.
SEMIA3007;M.
huakuii(CP006581.
1);M.
loti(NC_002678.
2);M.
opportunistum(NC_015675.
1);M.
erdmanii(NZ_AXAE01000048.
1)Kishietal.
BMCMicrobiology(2016)16:260Page6of8polymerase(LudwigBiotec).
AthermocyclermodelPTC-100ProgrammableThermalController(MJResearch,Inc.
)wasusedwithathermalprofileof96°Cfor2min,40cyclesof96°Cfor30s,53°Cfor1minand60°Cfor4min.
AfterthePCRreaction,theproductswerepurifiedwithaWizardSVGelandPCRClean-UpSystem(Pro-mega).
Theampliconwassequencedwith1μlofBigDyeTerminatorv3.
1,buffer0.
75X(Tris-HCl200mM,pH9.
0andMgCl25mM),10pmolesofprimerFD1,50ngofDNAandsterileMilli-Qdistilledwater(10μLq.
s.
p).
SequencingwasperformedonSequen-cerABIPRISM3130xlDNAAnalyzer(AppliedBiosystems)followingthemanufacturer'sinstructions.
Downloadingthesequences16SrRNAinGenBankTheNationalCenterforBiotechnologyInformation(NCBI)wasusedtosearchthegenomeforspeciesMesorhizobium(March15,2016).
Allcompletegenese-quencesfor16SrRNA(16SribosomalRNA)weredown-loadedfromGenBank(Additionalfile1:TableS1)[46].
Phylogeneticanalysisof16SrRNAgeneThe16SrRNAgenesetwerealignedusingtheMAFFTv7.
215program[47].
ThesearchforthebestnucleotidesubstitutionmatrixwasperformedwiththePhangornpackage[48]inR[49]andthefeaturemodelTest.
TheconstructionofaphylogenetictreewasperformedwiththeMrbayesv3.
2.
2program[50]usingthematrixreplacementGeneralTimeReversible(GTR)withgammavariation(G)andinvariablesites(I)with10.
000.
000generations.
ThebestevolutionarymodelwaschosenbasedonAkaikeinformationcriterionwithcorrection(AICc).
NucleotidesequenceaccessionnumberThedatasetsresultsofthisarticleareavailableintheNCBIBioProjectSRR3703040.
AdditionalfileAdditionalfile1:TableS1.
Completegenomesanddrafts.
(XLSX18kb)Abbreviations16SrRNA:16SribosomalRNAgene;AICc:Akaikeinformationcriterionwithcorrection;ANI:Averagenucleotideidentityvalues;CDS:Codingsequences;DDH:DNA-DNAhybridization;DNA:Deoxyribonucleicacid;GTR:Generaltimereversible;LCB:Collinearblocksconserved;LCB:Locallycollinearblocks;NAD(P)H:Nitritereductase;NADH:Glutamatesynthase;NADPH:Glutamatesynthase;NCBI:NationalCenterforBiotechnologyInformation;NGS:Nextgenerationsequencing;OD:Opticaldensity;ORF:Openreadframe;PCR:Polymerasechainreaction;TY:Triptone,yeastmediumAcknowledgementsWegratefullyacknowledgeUniversidadeEstadualPaulista–UNESP,theProgramadePósGraduaoemMicrobiologiaAgropecuária,UNESP,Jaboticabal,SoPauloState,Brazil.
WealsothankFAPESP(grants:2009/539842;2014/14234-6)andCNPq.
FundingThisworkwassupportedbyFAPESP(grants:2009/539842;2014/14234-6)andCNPq.
AvailabilityofdataandmaterialsThedatasetsresultsofthisarticleareavailableintheNCBIBioProjectSRR3703040.
http://www.
ncbi.
nlm.
nih.
gov/sra/term=SRR3703040Thedatasetsresultsfromphylogenetictreeofthisarticleareavaiablein:http://purl.
org/phylo/treebase/phylows/study/TB2:S20064YoucancitethisURLinyourmanuscript.
Itwillbecomethepermanentandresolvableresourcelocatorafteryoursubmissionhasbeenapprovedandthedataaremadepublic.
http://purl.
org/phylo/treebase/phylows/study/TB2:S20064x-access-code=c3247bbe7382039c0d043d51a87de8e8&format=htmlYoucancopyandsendthisURLtoyoujournaleditortoprovidereviewerswithlimited,read-onlyaccesstoyourdata,evenifyoursubmissionhasnotyetbeenapprovedandthedataarenotyetpublic.
Authors'contributionsConceivedanddesignedtheexperiments:LTKCCFEGML.
Performedtheexperiments:LTKCCFJCCEML.
Analyzedthedata:LTKWPO.
Contributedreagents/materials/analysistools:EGML.
Wrotethepaper:CCFLTK.
Allauthorsreadandapprovedthefinalmanuscript.
CompetinginterestsTheauthorsdeclarethattheyhavenocompetinginterests.
ConsentforpublicationAllauthorsareawareofthepublication.
EthicsapprovalandconsenttoparticipateAllauthorsareawareofthepublication.
Received:29July2016Accepted:28October2016References1.
KanekoT,NakamuraY,SatoS,AsamizuE,KatoT,SasamotoS,etal.
Completegenomestructureofthenitrogen-fixingsymbioticbacteriumMesorhizobiumloti.
DNARes.
IntJRapidPublRepGenesGenomes.
2000;7:331–8.
2.
VelázquezE,PeixA,Zurdo-PiiroJL,PalomoJL,MateosPF,RivasR,etal.
Thecoexistenceofsymbiosisandpathogenicity-determininggenesinRhizobiumrhizogenesstrainsenablesthemtoinducenodulesandtumorsorhairyrootsinplants.
MolPlantMicrobeInteract.
2005;18:1325–32.
3.
GrahamPH,SadowskyMJ,KeyserHH,BarnetYM,BradleyRS,CooperJE,etal.
Proposedminimalstandardsforthedescriptionofnewgeneraandspeciesofroot-andstem-nodulatingbacteria.
IntJSystBacteriol.
1991;41:582–7.
4.
WangS,HaoB,LiJ,GuH,PengJ,XieF,etal.
Whole-genomesequencingofMesorhizobiumhuakuii7653Rprovidesmolecularinsightsintohostspecificityandsymbiosisislanddynamics.
BMCGenomics.
2014;15:440.
5.
DegefuT,Wolde-meskelE,LiuB,CleenwerckI,WillemsA,FrostegardA.
Mesorhizobiumshonensesp.
nov.
,Mesorhizobiumhawassensesp.
nov.
andMesorhizobiumabyssinicaesp.
nov.
,isolatedfromrootnodulesofdifferentagroforestrylegumetrees.
IntJSystEvolMicrobiol.
2013;63:1746–53.
6.
JarvisBDW,VanBerkumP,ChenWX,NourSM,FernandezMP,Cleyet-MarelJC,etal.
Transferofrhizobiumloti,rhizobiumhuakuii,rhizobiumciceri,rhizobiummediterraneum,andrhizobiumtianshanensetomesorhizobiumgen.
nov.
IntJSystBacteriol.
1997;47:895–8.
7.
MousaviSA,WillemsA,NesmeX,deLajudieP,LindstrmK.
Revisedphylogenyofrhizobiaceae:proposalofthedelineationofpararhizobiumgen.
nov.
,and13newspeciescombinations.
SystApplMicrobiol.
2015;38:84–90.
8.
MousaviSA,stermanJ,WahlbergN,NesmeX,LavireC,VialL,etal.
PhylogenyoftheRhizobium–Allorhizobium–AgrobacteriumcladesupportsthedelineationofNeorhizobiumgen.
nov.
SystApplMicrobiol.
2014;37:208–15.
9.
Martínez-HidalgoP,Martínez-MolinaE,MateosPF,VelázquezE,Peix,Flores-FélixJD,etal.
RevisionofthetaxonomicstatusoftypestrainsofKishietal.
BMCMicrobiology(2016)16:260Page7of8MesorhizobiumlotiandreclassificationofstrainUSDA3471TasthetypestrainofMesorhizobiumerdmaniisp.
nov.
andATCC33669TasthetypestrainofMesorhizobiumjarvisiisp.
nov.
Int.
J.
Syst.
Evol.
Microbiol.
2015;65:1703–8.
10.
Ormeo-OrrilloE,Servín-GarcidueasLE,RogelMA,GonzálezV,PeraltaH,MoraJ,etal.
TaxonomyofrhizobiaandagrobacteriafromtheRhizobiaceaefamilyinlightofgenomics.
SystApplMicrobiol.
2015;38:287–91.
11.
GuptaA,SharmaVK.
Usingthetaxon-specificgenesforthetaxonomicclassificationofbacterialgenomes.
BMCGenomics[Internet].
2015[cited2016Oct4];16.
Availablefrom:http://www.
biomedcentral.
com/1471-2164/16/396.
Accessed5Oct2016.
12.
TengJLL,TangY,HuangY,GuoF-B,WeiW,ChenJHK,etal.
PhylogenomicAnalysesandReclassificationofSpecieswithintheGenusTsukamurella:InsightstoSpeciesDefinitioninthePost-genomicEra.
Front.
Microbiol.
[Internet].
2016[cited2016Oct4];7.
Availablefrom:http://journal.
frontiersin.
org/Article/10.
3389/fmicb.
2016.
01137/abstract.
Accessed5Oct2016.
13.
McIlroySJ,LapidusA,ThomsenTR,HanJ,HaynesM,LobosE,etal.
HighqualitydraftgenomesequenceofMeganemaperideroedesstr.
Gr1TandaproposalforitsreclassificationtothefamilyMeganemaceaefam.
nov.
Stand.
GenomicSci.
[Internet].
2015[cited2016Oct4];10.
Availablefrom:http://www.
standardsingenomics.
com/content/10/1/23.
Accessed5Oct2016.
14.
VandammeP,PotB,GillisM,deVosP,KerstersK,SwingsJ.
Polyphasictaxonomy,aconsensusapproachtobacterialsystematics.
MicrobiolRev.
1996;60:407–38.
15.
KimB-Y,WeonH-Y,CousinS,YooS-H,KwonS-W,GoS-J,etal.
Flavobacteriumdaejeonensesp.
nov.
andFlavobacteriumsuncheonensesp.
nov.
,isolatedfromgreenhousesoilsinKorea.
IntJSystEvolMicrobiol.
2006;56:1645–9.
16.
MennaP,HungriaM,BarcellosFG,BangelEV,HessPN,Martínez-RomeroE.
Molecularphylogenybasedonthe16SrRNAgeneofeliterhizobialstrainsusedinBraziliancommercialinoculants.
SystApplMicrobiol.
2006;29:315–32.
17.
RichterM,Rossello-MoraR.
Shiftingthegenomicgoldstandardfortheprokaryoticspeciesdefinition.
ProcNatlAcadSci.
2009;106:19126–31.
18.
CoenyeT,GeversD,VandePeerY,VandammeP,SwingsJ.
Towardsaprokaryoticgenomictaxonomy.
FEMSMicrobiolRev.
2005;29:147–67.
19.
KimM,LeeK-H,YoonS-W,KimB-S,ChunJ,YiH.
Analyticaltoolsanddatabasesformetagenomicsinthenext-generationsequencingera.
GenomicsInform.
2013;11:102.
20.
AchtmanM,WagnerM.
Microbialdiversityandthegeneticnatureofmicrobialspecies.
Nat.
Rev.
Microbiol.
[Internet].
2008[cited2016Jun22];Availablefrom:http://www.
nature.
com/nrmicro/journal/v6/n6/abs/nrmicro1872.
html.
Accessed5Oct2016.
21.
KonstantinidisKT,TiedjeJM.
Genomicinsightsthatadvancethespeciesdefinitionforprokaryotes.
ProcNatlAcadSci.
2005;102:2567–72.
22.
KonstantinidisKT,TiedjeJM.
Trendsbetweengenecontentandgenomesizeinprokaryoticspecieswithlargergenomes.
ProcNatlAcadSciUSA.
2004;101:3160–5.
23.
GorisJ,KlappenbachJA,VandammeP,CoenyeT,KonstantinidisKT,TiedjeJM.
DNA–DNAhybridizationvaluesandtheirrelationshiptowhole-genomesequencesimilarities.
IntJSystEvolMicrobiol.
2007;57:81–91.
24.
KohlerS,FoulongneV,Ouahrani-BettacheS,BourgG,TeyssierJ,RamuzM,etal.
Nonlinearpartialdifferentialequationsandapplications:theanalysisoftheintramacrophagicvirulomeofBrucellasuisdecipherstheenvironmentencounteredbythepathogeninsidethemacrophagehostcell.
ProcNatlAcadSci.
2002;99:15711–6.
25.
HaineV,DozotM,DornandJ,LetessonJ-J,DeBolleX.
NnrAisrequiredforfullvirulenceandregulatesseveralbrucellamelitensisdenitrificationgenes.
JBacteriol.
2006;188:1615–9.
26.
HubberAM,SullivanJT,RonsonCW.
Symbiosis-inducedcascaderegulationoftheMesorhizobiumlotiR7AVirB/D4typeIVsecretionsystem.
MolPlantMicrobeInteract.
2007;20:255–61.
27.
SotoMJ,SanjuanJ,OlivaresJ.
Rhizobiaandplant-pathogenicbacteria:commoninfectionweapons.
Microbiology.
2006;152:3167–74.
28.
BoschiroliML,Ouahrani-BettacheS,FoulongneV,Michaux-CharachonS,BourgG,Allardet-ServentA,etal.
TypeIVsecretionandBrucellavirulence.
VetMicrobiol.
2002;90:341–8.
29.
GarrityGM,TrüperHG,WhitmanWB,GrimontPAD,NesmeX,FrederiksenW,etal.
Reportoftheadhoccommitteeforthere-evaluationofthespeciesdefinitioninbacteriology.
IntJSystEvolMicrobiol.
2002;52:1043–7.
30.
AltschulSF,GishW,MillerW,MyersEW,LipmanDJ.
Basiclocalalignmentsearchtool.
JMolBiol.
1990;215:403–10.
31.
Rosselló-MoraR,AmannR.
Thespeciesconceptforprokaryotes.
FEMSMicrobiolRev.
2001;25:39–67.
32.
DelcherAL,KasifS,FleischmannRD,PetersonJ,WhiteO,SalzbergSL.
Alignmentofwholegenomes.
NucleicAcidsRes.
1999;27:2369–76.
33.
TeelingH,MeyerdierksA,BauerM,AmannR,GlocknerFO.
Applicationoftetranucleotidefrequenciesfortheassignmentofgenomicfragments.
EnvironMicrobiol.
2004;6:938–47.
34.
SchuldesJ,RodriguezOrbegosoM,SchmeisserC,KrishnanHB,DanielR,StreitWR.
Completegenomesequenceofthebroad-host-rangestrainsinorhizobiumfrediiUSDA257.
JBacteriol.
2012;194:4483.
35.
FitchWM.
Distinguishinghomologousfromanalogousproteins.
SystZool.
1970;19:99.
36.
LiL.
OrthoMCL:identificationoforthologgroupsforeukaryoticgenomes.
GenomeRes.
2003;13:2178–89.
37.
TatusovRL,KooninEV,LipmanDJ.
Agenomicperspectiveonproteinfamilies.
Science.
1997;278:631–7.
38.
ChervitzSA,AravindL,SherlockG,BallCA,KooninEV,DwightSS,etal.
Comparisonofthecompleteproteinsetsofwormandyeast:orthologyanddivergence.
Science.
1998;282:2022–8.
39.
BeringerJE.
RfactortransferinRhizobiumleguminosarum.
JGenMicrobiol.
1974;84:188–98.
40.
MartinM.
Cutadaptremovesadaptersequencesfromhigh-throughputsequencingreads.
EMBnetJ.
2011;17:10.
41.
SchmiederR,EdwardsR.
Qualitycontrolandpreprocessingofmetagenomicdatasets.
Bioinformatics.
2011;27:863–4.
42.
BankevichA,NurkS,AntipovD,GurevichAA,DvorkinM,KulikovAS,etal.
SPAdes:anewgenomeassemblyalgorithmanditsapplicationstosingle-cellsequencing.
JComputBiol.
2012;19:455–77.
43.
AzizRK,BartelsD,BestAA,DeJonghM,DiszT,EdwardsRA,etal.
TheRASTserver:rapidannotationsusingsubsystemstechnology.
BMCGenomics.
2008;9:75.
44.
DarlingACE.
Mauve:multiplealignmentofconservedgenomicsequencewithrearrangements.
GenomeRes.
2004;14:1394–403.
45.
WeisburgWG,BarnsSM,PelletierDA,LaneDJ.
16SribosomalDNAamplificationforphylogeneticstudy.
JBacteriol.
1991;173:697–703.
46.
LaranjoM,AlexandreA,OliveiraS.
Legumegrowth-promotingrhizobia:anoverviewontheMesorhizobiumgenus.
MicrobiolRes.
2014;169:2–17.
47.
KatohK.
MAFFT:anovelmethodforrapidmultiplesequencealignmentbasedonfastFouriertransform.
NucleicAcidsRes.
2002;30:3059–66.
48.
SchliepKP.
phangorn:phylogeneticanalysisinR.
Bioinformatics.
2011;27:592–3.
49.
IhakaR,GentlemanR.
R:Alanguagefordataanalysisandgraphics.
JComputGraphStat.
1995;5:299–314.
50.
RonquistF,TeslenkoM,vanderMarkP,AyresDL,DarlingA,HohnaS,etal.
MrBayes3.
2:efficientbayesianphylogeneticinferenceandmodelchoiceacrossalargemodelspace.
SystBiol.
2012;61:539–42.
Weacceptpre-submissioninquiriesOurselectortoolhelpsyoutondthemostrelevantjournalWeprovideroundtheclockcustomersupportConvenientonlinesubmissionThoroughpeerreviewInclusioninPubMedandallmajorindexingservicesMaximumvisibilityforyourresearchSubmityourmanuscriptatwww.
biomedcentral.
com/submitSubmityournextmanuscripttoBioMedCentralandwewillhelpyouateverystep:Kishietal.
BMCMicrobiology(2016)16:260Page8of8
- traits33669.com相关文档
- 募集33669.com
- medium33669.com
- LUCI33669.com
- 应答33669.com
- remarks33669.com
- 5.133669.com
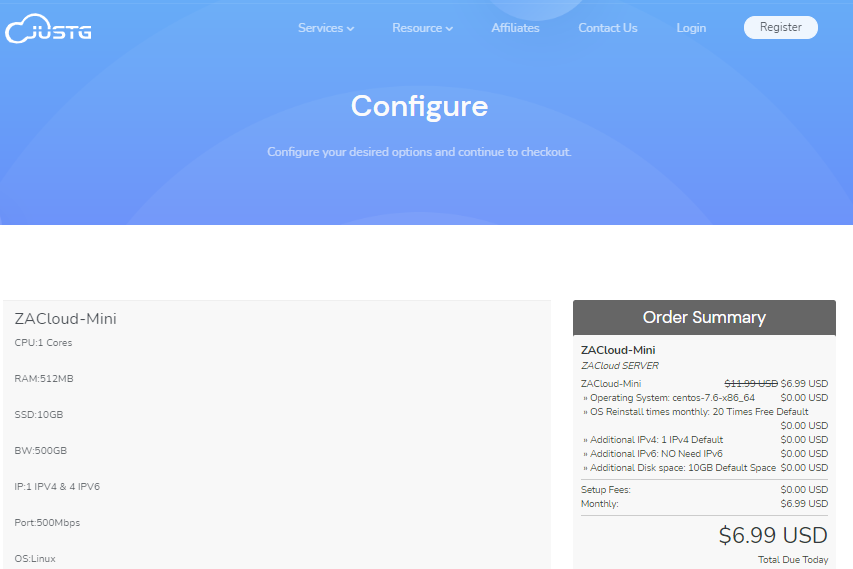
JUSTG(5.99美元/月)最新5折优惠,KVM虚拟虚拟512Mkvm路线
Justg是一家俄罗斯VPS云服务器提供商,主要提供南非地区的VPS服务器产品,CN2高质量线路网络,100Mbps带宽,自带一个IPv4和8个IPv6,线路质量还不错,主要是用户较少,带宽使用率不高,比较空闲,不拥挤,比较适合面向非洲、欧美的用户业务需求,也适合追求速度快又需要冷门的朋友。justg的俄罗斯VPS云服务器位于莫斯科机房,到美国和中国速度都非常不错,到欧洲的平均延迟时间为40毫秒,...
华纳云CN2高防1810M带宽独享,三网直cn218元/月,2M带宽;独服/高防6折购
华纳云怎么样?华纳云是香港老牌的IDC服务商,成立于2015年,主要提供中国香港/美国节点的服务器及网络安全产品、比如,香港服务器、香港云服务器、香港高防服务器、香港高防IP、美国云服务器、机柜出租以及云虚拟主机等。以极速 BGP 冗余网络、CN2 GIA 回国专线以及多年技能经验,帮助全球数十万家企业实现业务转型攀升。华纳云针对618返场活动,华纳云推出一系列热销产品活动,香港云服务器低至3折,...

QQ防红跳转短网址生成网站源码(91she完整源码)
使用此源码可以生成QQ自动跳转到浏览器的短链接,无视QQ报毒,任意网址均可生成。新版特色:全新界面,网站背景图采用Bing随机壁纸支持生成多种短链接兼容电脑和手机页面生成网址记录功能,域名黑名单功能网站后台可管理数据安装说明:由于此版本增加了记录和黑名单功能,所以用到了数据库。安装方法为修改config.php里面的数据库信息,导入install.sql到数据库。...

33669.com为你推荐
-
漏洞chrome朝阳分局电子物证实验室建设项目南京医科大学合同管理系统127.0.0.1为什么输入127.0.0.1无法打开页面x-routerX-TRAlL是什么意思ms17-010win10华为 slatl10是什么型号iphonewifi苹果手机怎么扫二维码连wifiicloudiphone苹果手机显示"已停用,连接itunes"是什么意思chromeframe我的Chrome Frame为什么不能使用?css选择器CSS中的选择器分几种?