Centerjpboy
jpboy 时间:2021-01-26 阅读:()
Iidaetal.
HumanGenomeVariation(2019)6:1https://doi.
org/10.
1038/s41439-018-0032-8HumanGenomeVariationDATAREPORTOpenAccessAnovelintragenicdeletioninOPHN1inaJapanesepatientwithDandy-WalkermalformationAritoshiIida1,EriTakeshita2,ShunichiKosugi3,YoichiroKamatani3,4,YukihideMomozawa5,MichiakiKubo6,EijiNakagawa2,KenjiKurosawa7,KenInoue8andYu-ichiGoto8,9AbstractDandy-Walkermalformation(DWM)isararecongenitalmalformationdenedbyhypoplasiaofthecerebellarvermisandcysticdilatationofthefourthventricle.
Oligophrenin-1ismutatedinX-linkedintellectualdisabilitywithorwithoutcerebellarhypoplasia.
Here,wereportaJapaneseDWMpatientcarryinganovelintragenic13.
5-kbdeletioninOPHN1rangingfromexon11–15.
ThisistherstreportofanOPHN1deletioninaJapanesepatientwithDWM.
Dandy-Walkermalformation(DWM)isamidbrain–hindbrainmalformationcharacterizedbycer-ebellarvermishypoplasiaanddysplasia,cysticdilatationofthefourthventricleandanelevatedtorcula,oftenaccompaniedbyhydrocephalus1.
ThefrequencyofDWMintheU.
S.
is~1in25,000–35,000liveborninfants(https://rarediseases.
org/rare-diseases/dandy-walker-malformation/).
DWMbecomesapparentinearlyinfancy,iscomplicatedbymacrocephaly,andoccursalongwithincreasedintracranialpressure,spasticparaparesis,andhypotonia2.
Inaddition,motordecits,suchasdelayedmotordevelopment,hypotonia,andataxia,aswellasintellectualdisability(ID),areoftenseen1,2.
Todate,variouschromosomalabnormalities,suchastrisomy9,13,18andpartialduplications/deletionsofchromo-somes,inDWMpatientshavebeenreviewed1.
Addi-tionally,heterozygousdeletionsofcerebellum-specicZinc-ngergenes,ZIC1andZIC4,onchromosome3q24areassociatedwithDWM3.
X-linkedDWMwithIDisalsocausedbyanAP1S2mutation4.
OPHN1encodesoligophrenin1,whichisaRho-GTPaseactivatingproteininvolvedinsynapticmorphogenesisandfunctionsthroughtheregulationoftheGproteincycle5.
OPHN1(NM_002547)consistsof25exonsandspans~391kbonchromosomeXq12(UCSCGenomeBrowser:https://genome.
ucsc.
edu).
Oligophrein1isan802amino-acidproteinharboringmultipledomains,suchasaBARdomain,PHdomain,Rho-GAPdomain,andthreeproline-richsequences6.
OPHN1wasoriginallyidentiedasadisruptedgenebyatranslocationt(X;12)inafemalepatientwithmildmentalretardation7.
Todate,10pointmutations,foursplicingmutations,sixsmallinsertion/deletionorduplicationmutations,and17chromosomalrearrangementsinOPHN1havebeenidentiedinpatientswithneurodevelopmentaldisorders,includingcerebellarhypoplasia,intellectualdisability(ID),epilepsy,seizure,ataxiaandschizophrenia(HumanGeneMutationDatabase,Professional2018.
2).
Inaddition,Schwartzetal.
6recentlyexpandedtheclinicalspectrumofOPHN1-associatedphenotypesincomparisontothephenotypesdescribedinpreviousreports.
Moortgatetal.
8alsodescribedfourfamilieswithintellectualdisabilitywithoutcerebellarhypoplasia.
Here,wereportaJapaneseDWMboycarryinganintragenicdeletioninOPHN1.
TheAuthor(s)2018OpenAccessThisarticleislicensedunderaCreativeCommonsAttribution4.
0InternationalLicense,whichpermitsuse,sharing,adaptation,distributionandreproductioninanymediumorformat,aslongasyougiveappropriatecredittotheoriginalauthor(s)andthesource,providealinktotheCreativeCommonslicense,andindicateifchangesweremade.
Theimagesorotherthirdpartymaterialinthisarticleareincludedinthearticle'sCreativeCommonslicense,unlessindicatedotherwiseinacreditlinetothematerial.
Ifmaterialisnotincludedinthearticle'sCreativeCommonslicenseandyourintendeduseisnotpermittedbystatutoryregulationorexceedsthepermitteduse,youwillneedtoobtainpermissiondirectlyfromthecopyrightholder.
Toviewacopyofthislicense,visithttp://creativecommons.
org/licenses/by/4.
0/.
Correspondence:Yu-ichiGoto(goto@ncnp.
go.
jp)1DepartmentofClinicalGenomeAnalysis,MedicalGenomeCenter,NationalCenterofNeurologyandPsychiatry(NCNP),Kodaira,Tokyo187-8551,Japan2DepartmentofChildNeurology,NationalCenterHospital,NCNP,Kodaira,Tokyo187-8551,JapanFulllistofauthorinformationisavailableattheendofthearticle.
OfcialjournaloftheJapanSocietyofHumanGenetics1234567890():,;1234567890():,;1234567890():,;1234567890():,;ThebiobankattheNationalCenterofNeurologyandPsychiatry(NCNP)isauniquebiorepositoryforneu-ropsychiatric,muscular,anddevelopmentaldiseasesinJapan(https://ncbiobank.
org/index-e.
html).
WecollectedDNAsamples,clinicalinformation,andcelllines,withinformedconsent,from583familieswithneurodevelop-mentaldiseasesthatwerediagnosedbetween2004and2016.
ThestudywasapprovedbytheethicalcommitteeofNCNP.
Thecelllinesweredevelopedbytheimmortali-zationofperipherallymphocyteswithEpstein-Barrvirus.
Weconductedthecandidategeneapproachonchro-mosomeXtoidentifythecausativegeneinthispatient.
Thefollowing19knowncausativegenesforXLIDwereanalyzedbyrepeatexpansionanalyses(FMR1andFMR2)andSangersequencing(PQBP1,ARX,MECP2,ATRX,RPS6KA3,IL1RAPL1,TM4SF2,PAK3,FACL4,OPHN1,AGTR2,ARHGEF6,GDI1,SLC6A8,FTSJ1,ZNF41,andDLG3).
WeperformeddirectsequencingofPCRampli-consusinganABI3730capillarysequencer(ThermoFisherScientic,Waltham,MA,USA)accordingtothestandardprotocol.
Wedeterminedthebreakpointbycomparingthesequenceofthepatientwithsequencesfromanunaffectedperson.
Thepatient(III-1)wasa2-year-oldboy,andhewasreferredfordevelopmentaldelayattheageof11months.
Hismaternalunclewasaffectedwithhydrocephalus(Fig.
1a).
Theboywasbornthroughnormaldeliverywithoutasphyxiaat39weeksofgestation.
Hisweightandheadcircumferencewere2756g(0.
6SD)and32cm(0.
9SD),respectively.
Heacquiredheadcontrolat8months,buthecouldnotsitaloneorspeakwords.
Braincomputedtomography(CT)andbrainmagneticreso-nanceimaging(MRI)suggestedDWM(Fig.
1b).
Whenhewas1yearold,hereceivedshuntingforhishydro-cephalus.
Attheageof1yearand3months,hisheight,weight,andheadcircumferencewere81cm(+1.
1SD),10.
7kg(+0.
8SD),and47cm(0SD),respectively,andhisdevelopmentalquotient(DQ)was57.
AfterinitialscreeningforanL1CAMmutationbysequencingandagrosscopynumbervariationinchro-mosomeXbyaBAC-basedarray-CGH9,whichwerebothnegative,weperformedmutationscreeningof19knowncausativegenesforXLIDinthepatient.
Consequently,wefoundanintragenicdeletioninOPHN1involvingexon11–15,whichincludethePHandGAPdomains(Fig.
2a).
Tomorepreciselydeterminethemechanismofthedeletion,weperformeddeletionmappingbyPCR-basedsequence-taggedsitecontentmapping,followedbydirectsequencingofthejunctionfragment.
Wenarrowedthebreakpointregiontoa437-bpPCRproductampliedbyasetofPCRprimersderivedfromintron10andintron15(SupplementaryFig.
1).
DirectsequencingofthePCRproductcontainingtherecombinationbreakpointrevealedthatthedeletiononlyoccurredwithinacommonve-nucleotidemotif(AATTA)inintron10andintron15,bothinthepatientandhismother.
Nolowcopyrepeatsorsegmentalduplicationswerefoundadjacenttothedeletionbreakpoints,suggestingthatthegenomicrearrangementoccurredbyamicrohomology-mediatedmechanismandnotbynon-allelichomologousrecombi-nation(Fig.
2b).
Thedeletionspannedfrom4218nucleotidesdownstreamoftheexon10-donorsite(c.
933+4,218)to4081nucleotidesdownstreamofexon15(c.
1276+4,081).
Thesizeofthedeletionwas13,517bpinlength(GRch37/hg19:chrX:67,408,680-67,422,196).
Thisdeletionwasalsoabsentinthreepublicdatabases,dbVar,ClinVarandtheDatabaseofGenomicVariants(http://dgv.
tcag.
ca/dgv/app/home).
Inaddition,wedidnotndthedeletionina1254Japanesegeneralpopulationdatasetcreatedbyhigh-depthwholegenomesequencing10.
Inpaststudies,OPHN1mutationshavebeenreportedincaseswithXLIDwithcerebellarhypoplasia,strabismus,epilepsy,hypotonia,ventriculomegaly,anddistinctiveFig.
1Thepedigreeandthebrainimages.
aPedigreeofthefamily.
Arrowindicatestheproband.
bBraincomputedtomographyimages(Upper)andaT2-weightedmagneticresonanceimage(Lower)ofthepatient.
Prominentcerebellarhypoplasia,anenlargedfourthventricleandposteriorfossa,andventriculomegalywerenotedIidaetal.
HumanGenomeVariation(2019)6:1Page2of4OfcialjournaloftheJapanSocietyofHumanGeneticsfacialfeatures6,8.
Additionally,OPHN1mutationshavealsobeenreportedinindividualswithautismorchildhoodonsetschizophrenia11,soOPHN1-associatedclinicalphenotypesarevariable6–8,11.
DWMandhydro-cephalusinthepresentpatientarelikelythemostsevereimagingndingsobservedinpatientswithOPHN1mutations.
ThenovelintragenicdeletioninOPHN1eliminatedexon11–15,whichencodePHandGAPdomains.
Thisdeletionleadstoaprematuretruncation,c.
934_1276del(p.
Gly312Ilefs*24),ofOPHN1;thetran-scriptmightpresumablybedegradedbynonsense-mediatedmRNAdecay.
Altogether,weconcludedthatthedeletioninOPHN1isthepathogenicgeneticabnormalityinthispatientwhoshowedprofoundIDandDWM.
HGVdatabaseTherelevantdatafromthisDataReportarehostedattheHumanGenomeVariationDatabaseathttps://doi.
org/10.
6084/m9.
gshare.
hgv.
2405AcknowledgementsWeareverygratefultothefamilywhoparticipatedinthisstudy.
WethankYoshieSawanoandShokoWatanabefortheirtechnicalassistance.
WealsothankDr.
JohjiInazawafortheBAC-basedarray-CGHanalysis.
ThisstudyispartiallysupportedbytheProgramforanIntegratedDatabaseofClinicalandGenomicInformation(17kk0205012h0002toY.
Goto),theConstructionofintegrateddatabaseofclinicalandgenomicsinformation,andthesustainablesystemforpromotinggenomicmedicineinJapan(18kk0205012s0303toY.
Goto)fromtheJapanAgencyforMedicalResearchandDevelopment,AMED,andtheIntramuralResearchGrant(27-6toY.
Goto;30-9toA.
Iida)forNeurologicalandPsychiatricDisordersofNCNP.
Authordetails1DepartmentofClinicalGenomeAnalysis,MedicalGenomeCenter,NationalCenterofNeurologyandPsychiatry(NCNP),Kodaira,Tokyo187-8551,Japan.
2DepartmentofChildNeurology,NationalCenterHospital,NCNP,Kodaira,Tokyo187-8551,Japan.
3LaboratoryforStatisticalAnalysis,RIKENCenterforIntegrativeMedicalSciences,Yokohama230-0045,Japan.
4Kyoto-McGillInternationalCollaborativeSchoolinGenomicMedicine,KyotoUniversityGraduateSchoolofMedicine,Kyoto606-8507,Japan.
5LaboratoryforGenotypingdevelopment,RIKENCenterforIntegrativeMedicalSciences,Yokohama230-0045,Japan.
6RIKENCenterforIntegrativeMedicalSciences,Yokohama230-0045,Japan.
7DivisionofMedicalGenetics,KanagawaChildren'sMedicalCenter,Yokohama,Kanagawa232-8555,Japan.
8DepartmentofMentalRetardationandBirthDefectResearch,NationalInstituteofNeuroscience,NCNP,Kodaira,Tokyo187-8551,Japan.
9DepartmentofBioresource,MedicalGenomeCenter,NCNP,Kodaira,Tokyo187-8551,JapanConictofinterestTheauthorsdeclarethattheyhavenoconictofinterest.
Publisher'snoteSpringerNatureremainsneutralwithregardtojurisdictionalclaimsinpublishedmapsandinstitutionalafliations.
Supplementaryinformationisavailableforthispaperathttps://doi.
org/10.
1038/s41439-018-0032-8.
Received:1October2018Revised:25October2018Accepted:2November2018References1.
Parisi,M.
A.
&Dobyns,W.
B.
Humanmalformationsofthemidbrainandhindbrain:reviewandproposedclassicationscheme.
Mol.
Genet.
Metab.
80,36–53(2003).
2.
Stambolliu,E.
,Ioakeim-Ioannidou,M.
,Kontokostas,K.
,Dakoutrou,M.
&Kou-soulis,A.
A.
ThemostcommoncomorbiditiesinDandy-Walkersyndromepatients:asystematicreviewofcasereports.
J.
ChildNeurol.
32,886–902(2017).
3.
Grinberg,I.
etal.
HeterozygousdeletionofthelinkedgenesZIC1andZIC4isinvolvedinDandy-Walkermalformation.
Nat.
Genet.
36,1053–1055(2004).
4.
Cacciagli,P.
etal.
AP1S2ismutatedinX-linkedDandy-Walkermalformationwithintellectualdisability,basalgangliadiseaseandseizures(Pettigrewsyn-drome).
Eur.
J.
Hum.
Genet.
22,363–368(2014).
5.
NadifKasri,N.
&VanAelst,L.
Rho-linkedgenesandneurologicaldisorders.
PugersArch.
455,787–797(2008).
6.
Schwartz,T.
S.
etal.
ExpandingthephenotypicspectrumassociatedwithOPHN1variants.
Eur.
J.
Med.
Genet.
S1769–7212,30005–3(2018).
7.
Billuart,P.
etal.
Oligophrenin-1encodesarhoGAPproteininvolvedinX-linkedmentalretardation.
Nature392,923–926(1998).
Fig.
2IdenticationofanintragenicdeletioninOPHN1inthepatientIII-1.
aAgarosegelimageofthePCRproductscorrespondingtointron10,exon12,andintron15inOPHN1inthepatientandhismother(II-2).
Exon12inthepatientwasdeleted.
ThegenomicstructureofOPHN1isalsoshownintheagarosegelimageofeachPCRanalysis.
PpositivecontrolDNAfromanunaffectedperson,Nnegativecontrol(H2O),M100-bpladderDNAsizemarker.
b(Upper)thedeletionmapofOPHN1.
Thegreenrectanglewithendbarsshowsthedeletedregion.
Thedeletionencompasses13.
5kbfromintron10tointron15.
(Lower)Theregionproximaltothedeletion,shownasaredbar,andtheregiondistaltothedeletion,shownasabluebar.
Sequencechromatogramsofthejunctionfragmentcontainingthebreakpointsitefromthepatientandhismotherareshowninthemiddletworows.
Anoverlapofave-nucleotidemotif(AATTA)isshownasapinkbar.
Referencesequencesofthecorrespondingregionsinintron10(upper)andintron15(lower)areshownIidaetal.
HumanGenomeVariation(2019)6:1Page3of4OfcialjournaloftheJapanSocietyofHumanGenetics8.
Moortgat,S.
etal.
ExpandingthephenotypicspectrumassociatedwithOPHN1mutations:reportof17individualswithintellectualdisabilitybutnocerebellarhypoplasia.
Eur.
J.
Med.
Genet.
S1769-7212,30666-3(2018).
9.
Hayashi,S.
etal.
Constructionofahigh-densityandhigh-resolutionhumanchromosomeXarrayforcomparativegenomichybridizationanalysis.
J.
Hum.
Genet.
52,397–405(2007).
10.
Okada,Y.
etal.
Deepwhole-genomesequencingrevealsrecentselectionsignatureslinkedtoevolutionanddiseaseriskofJapanese.
Nat.
Commun.
9,1631(2018).
11.
Piton,A.
etal.
SystematicresequencingofX-chromosomesynapticgenesinautismspectrumdisorderandschizopherinia.
Mol.
Psychiatry16,867–880(2011).
Iidaetal.
HumanGenomeVariation(2019)6:1Page4of4OfcialjournaloftheJapanSocietyofHumanGenetics
HumanGenomeVariation(2019)6:1https://doi.
org/10.
1038/s41439-018-0032-8HumanGenomeVariationDATAREPORTOpenAccessAnovelintragenicdeletioninOPHN1inaJapanesepatientwithDandy-WalkermalformationAritoshiIida1,EriTakeshita2,ShunichiKosugi3,YoichiroKamatani3,4,YukihideMomozawa5,MichiakiKubo6,EijiNakagawa2,KenjiKurosawa7,KenInoue8andYu-ichiGoto8,9AbstractDandy-Walkermalformation(DWM)isararecongenitalmalformationdenedbyhypoplasiaofthecerebellarvermisandcysticdilatationofthefourthventricle.
Oligophrenin-1ismutatedinX-linkedintellectualdisabilitywithorwithoutcerebellarhypoplasia.
Here,wereportaJapaneseDWMpatientcarryinganovelintragenic13.
5-kbdeletioninOPHN1rangingfromexon11–15.
ThisistherstreportofanOPHN1deletioninaJapanesepatientwithDWM.
Dandy-Walkermalformation(DWM)isamidbrain–hindbrainmalformationcharacterizedbycer-ebellarvermishypoplasiaanddysplasia,cysticdilatationofthefourthventricleandanelevatedtorcula,oftenaccompaniedbyhydrocephalus1.
ThefrequencyofDWMintheU.
S.
is~1in25,000–35,000liveborninfants(https://rarediseases.
org/rare-diseases/dandy-walker-malformation/).
DWMbecomesapparentinearlyinfancy,iscomplicatedbymacrocephaly,andoccursalongwithincreasedintracranialpressure,spasticparaparesis,andhypotonia2.
Inaddition,motordecits,suchasdelayedmotordevelopment,hypotonia,andataxia,aswellasintellectualdisability(ID),areoftenseen1,2.
Todate,variouschromosomalabnormalities,suchastrisomy9,13,18andpartialduplications/deletionsofchromo-somes,inDWMpatientshavebeenreviewed1.
Addi-tionally,heterozygousdeletionsofcerebellum-specicZinc-ngergenes,ZIC1andZIC4,onchromosome3q24areassociatedwithDWM3.
X-linkedDWMwithIDisalsocausedbyanAP1S2mutation4.
OPHN1encodesoligophrenin1,whichisaRho-GTPaseactivatingproteininvolvedinsynapticmorphogenesisandfunctionsthroughtheregulationoftheGproteincycle5.
OPHN1(NM_002547)consistsof25exonsandspans~391kbonchromosomeXq12(UCSCGenomeBrowser:https://genome.
ucsc.
edu).
Oligophrein1isan802amino-acidproteinharboringmultipledomains,suchasaBARdomain,PHdomain,Rho-GAPdomain,andthreeproline-richsequences6.
OPHN1wasoriginallyidentiedasadisruptedgenebyatranslocationt(X;12)inafemalepatientwithmildmentalretardation7.
Todate,10pointmutations,foursplicingmutations,sixsmallinsertion/deletionorduplicationmutations,and17chromosomalrearrangementsinOPHN1havebeenidentiedinpatientswithneurodevelopmentaldisorders,includingcerebellarhypoplasia,intellectualdisability(ID),epilepsy,seizure,ataxiaandschizophrenia(HumanGeneMutationDatabase,Professional2018.
2).
Inaddition,Schwartzetal.
6recentlyexpandedtheclinicalspectrumofOPHN1-associatedphenotypesincomparisontothephenotypesdescribedinpreviousreports.
Moortgatetal.
8alsodescribedfourfamilieswithintellectualdisabilitywithoutcerebellarhypoplasia.
Here,wereportaJapaneseDWMboycarryinganintragenicdeletioninOPHN1.
TheAuthor(s)2018OpenAccessThisarticleislicensedunderaCreativeCommonsAttribution4.
0InternationalLicense,whichpermitsuse,sharing,adaptation,distributionandreproductioninanymediumorformat,aslongasyougiveappropriatecredittotheoriginalauthor(s)andthesource,providealinktotheCreativeCommonslicense,andindicateifchangesweremade.
Theimagesorotherthirdpartymaterialinthisarticleareincludedinthearticle'sCreativeCommonslicense,unlessindicatedotherwiseinacreditlinetothematerial.
Ifmaterialisnotincludedinthearticle'sCreativeCommonslicenseandyourintendeduseisnotpermittedbystatutoryregulationorexceedsthepermitteduse,youwillneedtoobtainpermissiondirectlyfromthecopyrightholder.
Toviewacopyofthislicense,visithttp://creativecommons.
org/licenses/by/4.
0/.
Correspondence:Yu-ichiGoto(goto@ncnp.
go.
jp)1DepartmentofClinicalGenomeAnalysis,MedicalGenomeCenter,NationalCenterofNeurologyandPsychiatry(NCNP),Kodaira,Tokyo187-8551,Japan2DepartmentofChildNeurology,NationalCenterHospital,NCNP,Kodaira,Tokyo187-8551,JapanFulllistofauthorinformationisavailableattheendofthearticle.
OfcialjournaloftheJapanSocietyofHumanGenetics1234567890():,;1234567890():,;1234567890():,;1234567890():,;ThebiobankattheNationalCenterofNeurologyandPsychiatry(NCNP)isauniquebiorepositoryforneu-ropsychiatric,muscular,anddevelopmentaldiseasesinJapan(https://ncbiobank.
org/index-e.
html).
WecollectedDNAsamples,clinicalinformation,andcelllines,withinformedconsent,from583familieswithneurodevelop-mentaldiseasesthatwerediagnosedbetween2004and2016.
ThestudywasapprovedbytheethicalcommitteeofNCNP.
Thecelllinesweredevelopedbytheimmortali-zationofperipherallymphocyteswithEpstein-Barrvirus.
Weconductedthecandidategeneapproachonchro-mosomeXtoidentifythecausativegeneinthispatient.
Thefollowing19knowncausativegenesforXLIDwereanalyzedbyrepeatexpansionanalyses(FMR1andFMR2)andSangersequencing(PQBP1,ARX,MECP2,ATRX,RPS6KA3,IL1RAPL1,TM4SF2,PAK3,FACL4,OPHN1,AGTR2,ARHGEF6,GDI1,SLC6A8,FTSJ1,ZNF41,andDLG3).
WeperformeddirectsequencingofPCRampli-consusinganABI3730capillarysequencer(ThermoFisherScientic,Waltham,MA,USA)accordingtothestandardprotocol.
Wedeterminedthebreakpointbycomparingthesequenceofthepatientwithsequencesfromanunaffectedperson.
Thepatient(III-1)wasa2-year-oldboy,andhewasreferredfordevelopmentaldelayattheageof11months.
Hismaternalunclewasaffectedwithhydrocephalus(Fig.
1a).
Theboywasbornthroughnormaldeliverywithoutasphyxiaat39weeksofgestation.
Hisweightandheadcircumferencewere2756g(0.
6SD)and32cm(0.
9SD),respectively.
Heacquiredheadcontrolat8months,buthecouldnotsitaloneorspeakwords.
Braincomputedtomography(CT)andbrainmagneticreso-nanceimaging(MRI)suggestedDWM(Fig.
1b).
Whenhewas1yearold,hereceivedshuntingforhishydro-cephalus.
Attheageof1yearand3months,hisheight,weight,andheadcircumferencewere81cm(+1.
1SD),10.
7kg(+0.
8SD),and47cm(0SD),respectively,andhisdevelopmentalquotient(DQ)was57.
AfterinitialscreeningforanL1CAMmutationbysequencingandagrosscopynumbervariationinchro-mosomeXbyaBAC-basedarray-CGH9,whichwerebothnegative,weperformedmutationscreeningof19knowncausativegenesforXLIDinthepatient.
Consequently,wefoundanintragenicdeletioninOPHN1involvingexon11–15,whichincludethePHandGAPdomains(Fig.
2a).
Tomorepreciselydeterminethemechanismofthedeletion,weperformeddeletionmappingbyPCR-basedsequence-taggedsitecontentmapping,followedbydirectsequencingofthejunctionfragment.
Wenarrowedthebreakpointregiontoa437-bpPCRproductampliedbyasetofPCRprimersderivedfromintron10andintron15(SupplementaryFig.
1).
DirectsequencingofthePCRproductcontainingtherecombinationbreakpointrevealedthatthedeletiononlyoccurredwithinacommonve-nucleotidemotif(AATTA)inintron10andintron15,bothinthepatientandhismother.
Nolowcopyrepeatsorsegmentalduplicationswerefoundadjacenttothedeletionbreakpoints,suggestingthatthegenomicrearrangementoccurredbyamicrohomology-mediatedmechanismandnotbynon-allelichomologousrecombi-nation(Fig.
2b).
Thedeletionspannedfrom4218nucleotidesdownstreamoftheexon10-donorsite(c.
933+4,218)to4081nucleotidesdownstreamofexon15(c.
1276+4,081).
Thesizeofthedeletionwas13,517bpinlength(GRch37/hg19:chrX:67,408,680-67,422,196).
Thisdeletionwasalsoabsentinthreepublicdatabases,dbVar,ClinVarandtheDatabaseofGenomicVariants(http://dgv.
tcag.
ca/dgv/app/home).
Inaddition,wedidnotndthedeletionina1254Japanesegeneralpopulationdatasetcreatedbyhigh-depthwholegenomesequencing10.
Inpaststudies,OPHN1mutationshavebeenreportedincaseswithXLIDwithcerebellarhypoplasia,strabismus,epilepsy,hypotonia,ventriculomegaly,anddistinctiveFig.
1Thepedigreeandthebrainimages.
aPedigreeofthefamily.
Arrowindicatestheproband.
bBraincomputedtomographyimages(Upper)andaT2-weightedmagneticresonanceimage(Lower)ofthepatient.
Prominentcerebellarhypoplasia,anenlargedfourthventricleandposteriorfossa,andventriculomegalywerenotedIidaetal.
HumanGenomeVariation(2019)6:1Page2of4OfcialjournaloftheJapanSocietyofHumanGeneticsfacialfeatures6,8.
Additionally,OPHN1mutationshavealsobeenreportedinindividualswithautismorchildhoodonsetschizophrenia11,soOPHN1-associatedclinicalphenotypesarevariable6–8,11.
DWMandhydro-cephalusinthepresentpatientarelikelythemostsevereimagingndingsobservedinpatientswithOPHN1mutations.
ThenovelintragenicdeletioninOPHN1eliminatedexon11–15,whichencodePHandGAPdomains.
Thisdeletionleadstoaprematuretruncation,c.
934_1276del(p.
Gly312Ilefs*24),ofOPHN1;thetran-scriptmightpresumablybedegradedbynonsense-mediatedmRNAdecay.
Altogether,weconcludedthatthedeletioninOPHN1isthepathogenicgeneticabnormalityinthispatientwhoshowedprofoundIDandDWM.
HGVdatabaseTherelevantdatafromthisDataReportarehostedattheHumanGenomeVariationDatabaseathttps://doi.
org/10.
6084/m9.
gshare.
hgv.
2405AcknowledgementsWeareverygratefultothefamilywhoparticipatedinthisstudy.
WethankYoshieSawanoandShokoWatanabefortheirtechnicalassistance.
WealsothankDr.
JohjiInazawafortheBAC-basedarray-CGHanalysis.
ThisstudyispartiallysupportedbytheProgramforanIntegratedDatabaseofClinicalandGenomicInformation(17kk0205012h0002toY.
Goto),theConstructionofintegrateddatabaseofclinicalandgenomicsinformation,andthesustainablesystemforpromotinggenomicmedicineinJapan(18kk0205012s0303toY.
Goto)fromtheJapanAgencyforMedicalResearchandDevelopment,AMED,andtheIntramuralResearchGrant(27-6toY.
Goto;30-9toA.
Iida)forNeurologicalandPsychiatricDisordersofNCNP.
Authordetails1DepartmentofClinicalGenomeAnalysis,MedicalGenomeCenter,NationalCenterofNeurologyandPsychiatry(NCNP),Kodaira,Tokyo187-8551,Japan.
2DepartmentofChildNeurology,NationalCenterHospital,NCNP,Kodaira,Tokyo187-8551,Japan.
3LaboratoryforStatisticalAnalysis,RIKENCenterforIntegrativeMedicalSciences,Yokohama230-0045,Japan.
4Kyoto-McGillInternationalCollaborativeSchoolinGenomicMedicine,KyotoUniversityGraduateSchoolofMedicine,Kyoto606-8507,Japan.
5LaboratoryforGenotypingdevelopment,RIKENCenterforIntegrativeMedicalSciences,Yokohama230-0045,Japan.
6RIKENCenterforIntegrativeMedicalSciences,Yokohama230-0045,Japan.
7DivisionofMedicalGenetics,KanagawaChildren'sMedicalCenter,Yokohama,Kanagawa232-8555,Japan.
8DepartmentofMentalRetardationandBirthDefectResearch,NationalInstituteofNeuroscience,NCNP,Kodaira,Tokyo187-8551,Japan.
9DepartmentofBioresource,MedicalGenomeCenter,NCNP,Kodaira,Tokyo187-8551,JapanConictofinterestTheauthorsdeclarethattheyhavenoconictofinterest.
Publisher'snoteSpringerNatureremainsneutralwithregardtojurisdictionalclaimsinpublishedmapsandinstitutionalafliations.
Supplementaryinformationisavailableforthispaperathttps://doi.
org/10.
1038/s41439-018-0032-8.
Received:1October2018Revised:25October2018Accepted:2November2018References1.
Parisi,M.
A.
&Dobyns,W.
B.
Humanmalformationsofthemidbrainandhindbrain:reviewandproposedclassicationscheme.
Mol.
Genet.
Metab.
80,36–53(2003).
2.
Stambolliu,E.
,Ioakeim-Ioannidou,M.
,Kontokostas,K.
,Dakoutrou,M.
&Kou-soulis,A.
A.
ThemostcommoncomorbiditiesinDandy-Walkersyndromepatients:asystematicreviewofcasereports.
J.
ChildNeurol.
32,886–902(2017).
3.
Grinberg,I.
etal.
HeterozygousdeletionofthelinkedgenesZIC1andZIC4isinvolvedinDandy-Walkermalformation.
Nat.
Genet.
36,1053–1055(2004).
4.
Cacciagli,P.
etal.
AP1S2ismutatedinX-linkedDandy-Walkermalformationwithintellectualdisability,basalgangliadiseaseandseizures(Pettigrewsyn-drome).
Eur.
J.
Hum.
Genet.
22,363–368(2014).
5.
NadifKasri,N.
&VanAelst,L.
Rho-linkedgenesandneurologicaldisorders.
PugersArch.
455,787–797(2008).
6.
Schwartz,T.
S.
etal.
ExpandingthephenotypicspectrumassociatedwithOPHN1variants.
Eur.
J.
Med.
Genet.
S1769–7212,30005–3(2018).
7.
Billuart,P.
etal.
Oligophrenin-1encodesarhoGAPproteininvolvedinX-linkedmentalretardation.
Nature392,923–926(1998).
Fig.
2IdenticationofanintragenicdeletioninOPHN1inthepatientIII-1.
aAgarosegelimageofthePCRproductscorrespondingtointron10,exon12,andintron15inOPHN1inthepatientandhismother(II-2).
Exon12inthepatientwasdeleted.
ThegenomicstructureofOPHN1isalsoshownintheagarosegelimageofeachPCRanalysis.
PpositivecontrolDNAfromanunaffectedperson,Nnegativecontrol(H2O),M100-bpladderDNAsizemarker.
b(Upper)thedeletionmapofOPHN1.
Thegreenrectanglewithendbarsshowsthedeletedregion.
Thedeletionencompasses13.
5kbfromintron10tointron15.
(Lower)Theregionproximaltothedeletion,shownasaredbar,andtheregiondistaltothedeletion,shownasabluebar.
Sequencechromatogramsofthejunctionfragmentcontainingthebreakpointsitefromthepatientandhismotherareshowninthemiddletworows.
Anoverlapofave-nucleotidemotif(AATTA)isshownasapinkbar.
Referencesequencesofthecorrespondingregionsinintron10(upper)andintron15(lower)areshownIidaetal.
HumanGenomeVariation(2019)6:1Page3of4OfcialjournaloftheJapanSocietyofHumanGenetics8.
Moortgat,S.
etal.
ExpandingthephenotypicspectrumassociatedwithOPHN1mutations:reportof17individualswithintellectualdisabilitybutnocerebellarhypoplasia.
Eur.
J.
Med.
Genet.
S1769-7212,30666-3(2018).
9.
Hayashi,S.
etal.
Constructionofahigh-densityandhigh-resolutionhumanchromosomeXarrayforcomparativegenomichybridizationanalysis.
J.
Hum.
Genet.
52,397–405(2007).
10.
Okada,Y.
etal.
Deepwhole-genomesequencingrevealsrecentselectionsignatureslinkedtoevolutionanddiseaseriskofJapanese.
Nat.
Commun.
9,1631(2018).
11.
Piton,A.
etal.
SystematicresequencingofX-chromosomesynapticgenesinautismspectrumdisorderandschizopherinia.
Mol.
Psychiatry16,867–880(2011).
Iidaetal.
HumanGenomeVariation(2019)6:1Page4of4OfcialjournaloftheJapanSocietyofHumanGenetics
云基Yunbase无视CC攻击(最高500G DDoS防御),美国洛杉矶CN2-GIA高防独立服务器,
云基yunbase怎么样?云基成立于2020年,目前主要提供高防海内外独立服务器,欢迎各类追求稳定和高防优质线路的用户。业务可选:洛杉矶CN2-GIA+高防(默认500G高防)、洛杉矶CN2-GIA(默认带50Gbps防御)、香港CN2-GIA高防(双向CN2GIA专线,突发带宽支持,15G-20G DDoS防御,无视CC)。目前,美国洛杉矶CN2-GIA高防独立服务器,8核16G,最高500G ...

域名注册需要哪些条件(新手注册域名考虑的问题)
今天下午遇到一个网友聊到他昨天新注册的一个域名,今天在去使用的时候发现域名居然不见。开始怀疑他昨天是否付款扣费,以及是否有实名认证过,毕竟我们在国内域名注册平台注册域名是需要实名认证的,大概3-5天内如果不验证那是不可以使用的。但是如果注册完毕的域名找不到那也是奇怪。同时我也有怀疑他是不是忘记记错账户。毕竟我们有很多朋友在某个商家注册很多账户,有时候自己都忘记是用哪个账户的。但是我们去找账户也不办...
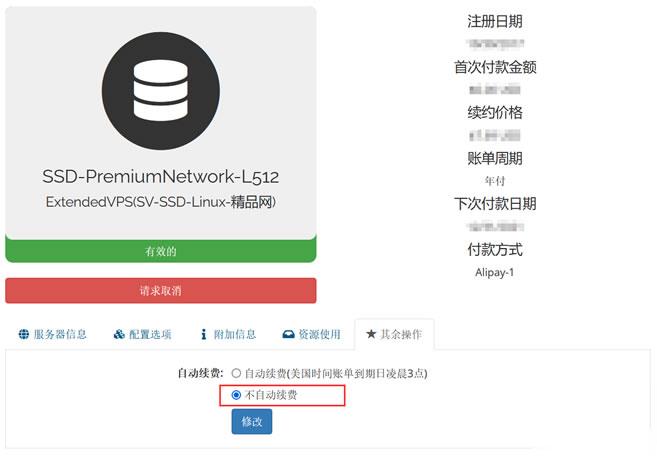
Raksmart VPS主机如何设置取消自动续费
今天有看到Raksmart账户中有一台VPS主机即将到期,这台机器之前是用来测试评测使用的。这里有不打算续费,这不面对万一导致被自动续费忘记,所以我还是取消自动续费设置。如果我们也有类似的问题,这里就演示截图设置Raksmart取消自动续费。这里我们可以看到上图,在对应VPS主机的【其余操作】中可以看到默认已经是不自动续费,所以我们也不要担心被自动续费的。当然,如果有被自动续费,我们确实不想续费的...
jpboy为你推荐
-
迈腾和帕萨特哪个好大众新帕萨特和迈腾哪个更好!小说软件哪个好用免费有什么好用的免费小说软件手机杀毒软件哪个好手机杀毒软件那个好用核芯显卡与独立显卡哪个好英特尔核芯显卡怎么样?和独立显卡那个更好?雅思和托福哪个好考托福好考还是雅思好考哇?qq空间登录器QQ空间校友网页自动登陆器qq空间登录器怎样直接登录QQ空间51个人空间登录51.com个人空间怎么无法登录?qq空间登录不上为什么我的qq空间登不上去qq空间登录电脑手机怎么登qq空间电脑版?