Microarray123408.com
123408.com 时间:2021-03-21 阅读:()
ArraycomparativegenomichybridizationanalysisofolfactoryneuroblastomaMohamedGuled1,SamuelMyllykangas1,HenryFFriersonJr2,StaceyEMills2,SakariKnuutila1andEdwardBStelow21DepartmentofPathology,HaartmanInstituteandHUSLAB,UniversityofHelsinkiandHelsinkiUniversityCentralHospital,Helsinki,Finlandand2DepartmentofPathology,UniversityofVirginiaHealthSystem,Charlottesville,VA,USAOlfactoryneuroblastomaisanunusualneuroectodermalmalignancy,whichisthoughttoariseattheolfactorymembraneofthesinonasaltract.
Duetoitsrarity,littleisunderstoodregardingitsmolecularandcytogeneticabnormalities.
TheaimofthecurrentstudyistoidentifyspecificDNAcopynumberchangesinolfactoryneuroblastoma.
Thirteendissectedtissuesampleswereanalyzedusingarraycomparativegenomichybridization.
Ourresultsshowthatgenecopynumberprofilesofolfactoryneuroblastomasamplesarecomplex.
Themostfrequentchangesincludedgainsat7q11.
22–q21.
11,9p13.
3,13q,20p/q,andXp/q,andlossesat2q31.
1,2q33.
3,2q37.
1,6q16.
3,6q21.
33,6q22.
1,22q11.
23,22q12.
1,andXp/q.
Gainsweremorefrequentthanlosses,andhigh-stagetumorsshowedmorealterationsthanlow-stageolfactoryneuroblastoma.
Frequentchangesinhigh-stagetumorsweregainsat13q14.
2–q14.
3,13q31.
1,and20q11.
21–q11.
23,andlossofXp21.
1(in66%ofcases).
Gainsat5q35,13q,and20q,andlossesat2q31.
1,2q33.
3,and6q16–q22,werepresentin50%ofcases.
Theidentifiedregionsofgenecopynumberchangehavebeenimplicatedinavarietyoftumors,especiallycarcinomas.
Inaddition,ourresultsindicatethatgainsin20qand13qmaybeimportantintheprogressionofthiscancer,andthattheseregionspossiblyharborgeneswithfunctionalrelevanceinolfactoryneuroblastoma.
ModernPathology(2008)21,770–778;doi:10.
1038/modpathol.
2008.
57;publishedonline11April2008Keywords:olfactoryneuroblastoma;arrayCGH;esthesioneuroblastoma;cytogeneticsOlfactoryneuroblastomaisrare,withanestimatedincidenceof0.
4permillionpeopleperyear.
Itaccountsforapproximately2–3%ofsinonasaltracttumors.
1Olfactoryneuroblastomaisamalignantneuroectodermaltumor,whichisbelievedtoorigi-nateattheolfactorymembraneofthesinonasaltract.
1Symptomsincludeunilateralnasalobstruc-tionandepistaxis.
TheKadishsystemclassifiescasesofolfactoryneuroblastomaintothreestages.
Stage1consistsoftumorsconfinedtothenasalcavity;stage2tumorsextendtotheparanasalsinuses;andstage3neoplasmsextendbeyondthesinonasalcavities(somehaveaddedastage4forthosetumorswithmetastases).
Patientswithhigh-stagetumorshaveaworseprognosiscomparedwiththosewithlow-stagetumors,as5-yearsurvivalratesare40and80%,respectively.
2Cytogeneticdataforolfactoryneuroblastomaarelimited.
ThemostrecentlypublishedstudywascarriedoutbyHollandetal,3whoperformedcytogeneticcharacterizationofonecaseusingtrypsinGiemsastaining(GTGbanding),multicolorfluorescenceinsituhybridization(M-FISH),andsingle-nucleotidepolymorphismkaryotyping.
Theyreportednumerouschromosomalaberrationspre-dominantlyinvolvingchromosomes2q,5,6q,17,19,21q,and22,aswellastrisomy8.
Bockmu¨ehletal4appliedconventionalcomparativegenomichy-bridization(CGH)to22olfactoryneuroblastomasandreportedfrequentdeletionsof1p,3p/q,9p,and10p/q,andamplificationsof17q,17p13,20p,and22q.
Theyalsonotedaspecificdeletiononchromo-some11andgainonchromosome1p,whichwereassociatedwithmetastasisandaworseprognosis.
ThreeolfactoryneuroblastomaswerestudiedbyRiazimandetal5usingconventionalCGH,andamplificationofwholechromosome19,partialgainsof1p,8q,15q,and22q,anddeletionsof4qand6pReceived24November2007;revised02March2008;accepted10March2008;publishedonline11April2008Correspondence:DrEBStelow,MD,DepartmentofPathology,UniversityofVirginiaHealthSciences,Box800214,JeffersonParkAvenue,Charlottesville,VA22908,USA.
E-mail:edstelow@yahoo.
comandDrSKnuutila,PhD,DepartmentofPathology,HaartmanInstituteandHUSLAB,POBox21(Haartmaninkatu3),UniversityofHelsinki,FI-00014Helsinki,Finland.
E-mail:Sakari.
Knuutila@helsinki.
fiModernPathology(2008)21,770–778&2008USCAP,IncAllrightsreserved0893-3952/08$30.
00www.
modernpathology.
orgweredetected.
Szymasetal6studiedasingleolfactoryneuroblastomaandfoundgainsofwholechromosomes4,8,11,and14,partialgainsof1qand17q,partialdeletionsof5qand17q,andwholechromosomelossesof16,18,19,andX.
Inourstudy,weappliedforthefirsttimeanoligonucleo-tide-basedarrayCGH(aCGH)toidentifythemostfrequentlyoccurringDNAcopynumberchangesin13casesofolfactoryneuroblastoma.
Weidentifiednovelchromosomalregionsthatwerefrequentlyalteredinadditiontopreviouslyreportedabnormalregions.
MaterialsandmethodsTumorSamplesTheSurgicalPathologyDatabase(DepartmentofPathologyattheUniversityofVirginia,Charlottes-ville,VA,USA)wassearchedforresectedcasesofolfactoryneuroblastoma.
Slidesfromformalin-fixed,paraffin-embeddedtissueswerereviewed,and15caseswereselectedbasedontheavailabilityofabundantandwell-preservedneoplastictissue.
Onlycaseswithconventionalhistology,asde-scribedbyMillsandFrierson,7werechosen(Figure1).
Allcasesexhibitedatypicalimmunophenotypeandwereimmunoreactivewithantibodiestosynap-tophysinandnotwithantibodiestokeratins.
Areasrepresentingcancertissueweredissectedfromparaffin-embeddedtumorsusingconventionallightmicroscopyandscalpelblades.
All15samplessubmittedforanalysiswerecomposedofatleast50%tumorcells.
EachofsixsampleswasclassifiedasKadishstage2and3(Figure1),whereasinformationregardingthestagewasnotavailableforthreetumors.
Patients'agerangedfrom27to62years.
TheclinicopathologicaldataforthepatientsandsamplesaresummarizedinTable1.
DNAExtractionandDigestionTotalDNAwasextractedusingQIAampDNAminikit,accordingtothemanufacturer'sinstructions(Qiagen,Hilden,Germany).
DNAconcentrationsoftheextractedsamplesweredeterminedusingtheNanoDropND-1000spectrophotometer(Nanodroptechnologies,Wilmington,DE,USA)at260/280nm.
TheintegrityoftheextractedDNAwasverifiedbyagarosegelelectrophoresis(datanotshown).
A1.
5mgquantityofgenomicDNAfromtestandreferencesampleswasdigestedusingAlu1andRsa1restrictionenzymes(Promega,Madison,WI,USA)for2hat371C.
Toinactivatetheenzymes,thesampleswereincubatedat651Cfor20min.
ArrayCGHExperimentsArrayCGHwasperformedfor13samplesthathadgoodqualityDNAwithsufficientyields.
ForFigure1StandardH&Esectionsofconventionalolfactoryneuroblastoma:(a)sample11(100),(b)sample12(200).
Table1ClinicopathologicaldataforthesamplesSampleSex/ageDead/aliveDiseasestatusTime(months)Stage(Kadish)1m/50DWD15622m/49ANED192UNK3m/53DWD2034f/38AUNK136UNK5f/40AUNK14926f/27ANED11937f/61ANED10638m/27ANED5929m/40ANED57312f/62DWD35313m/41UNKUNKUNK314m/44ANED24UNK15m/60ANED62Abbreviations:A,alive;D,dead;NED,noevidenceofdisease;UNK,unknown;WD,withdisease.
MicroarrayanalysisofolfactoryneuroblastomaMGuledetal771ModernPathology(2008)21,770–778reference,pooledDNAextractedfrombuffycoatfractionsofwholebloodobtainedfromsex-matched,healthyindividualsprovidedbytheFinnishRedCrosswasused.
TheAgilentgenomicDNAlabelingkitPLUS(AgilentTechnologies,SantaClara,CA,USA)wasusedtolabel1.
5mgofdigestedgenomicDNA.
TheDNAofolfactoryneuroblastomasamplesandthereferenceDNAwerelabeledwithcyanine5-dUTPandcyanine3-dUTPfluorochromes,respec-tively,for2hat371C.
Forinactivationoftheenzymes,thesampleswereincubatedat651Cfor10min.
Humancot-1DNA(Invitrogen,Carlsbad,CA,USA)wasaddedtothereactionsfor3minat951Candthenfor30minat371C.
Priortohybridiza-tionexperiments,labeledDNAwascleanedaccord-ingtoAgilent'sprotocol.
LabeledsampleandreferenceDNAwerehybridizedonAgilent's444KCGHmicroarray(productnumberG4426AandG4426B,AgilentTechnologies)for24hat651C.
Priortoscanning,microarrayslideswerewashedaccordingtothemanufacturer'sinstructions.
ThemicroarrayslideswerescannedusingDNAMicro-arrayScannerandthescannedimageswereana-lyzedusingtheFeatureExtractionSoftware(AgilentTechnologies).
DatawereimportedtotheCGHAnalyticsSoftware3.
4foranalysisofindividualcases.
Z-scorealgorithmwiththresholdat3.
5wasusedintheidentificationofcopynumberchanges.
Toobtainaglobalviewofthecopynumberalterationfrequenciesinolfactoryneuroblastoma,wealsoanalyzedthedatausingGeneSpring(AgilentTechnologies)andCGHExplorersoftware3.
1.
8TherawdatafromtheFeatureExtractionsoftwarewereimportedintotheGeneSpringsoftwareandnormal-izedusingtheFeatureExtractiondataimportplug-in(AgilentTechnologies).
OutlierprobesmarkedbyFeatureExtractionwereignoredinnormalization.
Samplespotintensitiesweredividedbycontrolchannelintensities,andmicroarrayswerenormal-izedtothe50thpercentileandprobestothemedianvalue.
Outlierprobeswerefilteredfromfurtheranalysis.
Normalizedandfiltereddatawereim-portedintotheCGHExplorersoftware.
CopynumberaberrationswereidentifiedusingPiecewiseConstantFit(PCF)algorithm.
Defaultparameterswereappliedtodetectlongcopynumberaberra-tions.
Thesignificancethresholdwassetat0.
2.
ResultsGeneCopyNumberProfilingofOlfactoryNeuroblastomaArrayCGHwasappliedtostudygenecopynumberalterationsin13olfactoryneuroblastomasamples.
TheCGHExplorersoftwarewasusedtoanalyzeglobalfrequenciesofcopynumberchange(Figure2).
Atotalof15.
74%oftheprobeswerefoundaberrantwhenathresholdof0.
2wasusedinthePCFanalysis.
DNAcopynumbergains(8.
37%)wereslightlymorefrequentthanlosses(7.
36%).
Inaddition,theCGHanalyticssoftwarewasusedtodeterminecopynumberaberrationsinindividualsamples.
CopynumberaberrationsforeachcasearesummarizedinTable2.
Changesinthecopynumberofentirechromosomes(aneuploidy)orchromosome12345678910111213141516171819202122XY0255075100255075100AberrationfrequencyGenomicpositionGainsLossesFigure2Frequencyprofileofcopynumberaberrationsin13olfactoryneuroblastomasamples.
CGHExplorersoftwareandPiecewiseConstantFitalgorithmwereusedtodeterminecopynumberaberrationsinolfactoryneuroblastomasamples.
Thechromosomalalterationsareshownineachprobepositionasincidencebar.
Gainsofgenomicmaterialareindicatedingrayontheuppersideofthemiddleline(at0).
Lossesareindicatedinblackonthebottomsideofthemiddleline.
GenomicpositionsoftheaCGHprobesaremarkedonthexaxis.
MicroarrayanalysisofolfactoryneuroblastomaMGuledetal772ModernPathology(2008)21,770–778armsrepresented16%ofallalterations;interest-ingly,themostalteredwholechromosomeswere20,21,22,andX,andalmost70%oftheaberrationsofthesewerewholechromosomelossesorgains.
Toobtainamoredetailedoverviewofthemostfrequentlyoccurringalterations,onlythosealtera-tionssharedbyatleast20%ofthecaseswererecorded.
Thisapproachidentifiedanequalnumberofgainedandlostchromosomalregions(n21).
Tables3and4showthesizeofthealteredareasandwhethertheyhavebeenreportedinpreviouscopynumberchangestudiesforolfactoryneuroblastoma.
Thetablesalsoincludesuggestedtargetgenesthatarelocatedatthesitesofalterationandthatmayplayaroleintumorigenesis.
SpecificCopyNumberAlterationsWereIdentifiedinCasesofOlfactoryNeuroblastomaofDifferentStageAstage3olfactoryneuroblastomashowedthemostDNAcopynumberchanges,with59%ofallalterations.
Also,mostaneuploidyoccurredinhigh-stagetumors;70%ofwhole-chromosomeaber-rationswereidentifiedinthesetumors.
Forstage3tumors,themeanDNAcopynumberchangepercasewas28.
5,whereasstage2olfactoryneuroblas-tomashadameannumberof17changespertumor.
However,only4outof6stage2casescontainedsufficientgoodqualityDNAtoperformaCGH.
Tofurthercharacterizethegenomicchangesthatoccurredinalater-stageolfactoryneuroblastoma,Table2aCGHresultsfor13ONBsamplesONBsampleStageTumorDNAcopynumberchanges12+1p31.
2,+1p31.
1,+1p35.
3,+1q31.
1q31.
3,+2p24.
3,2p25.
1p25.
3,2q36.
3q37.
3,+3p13.
12p13.
31,+4p,+4q,+5q23.
1,+8q22.
3q24.
12,9p21.
1,10p12.
31,11q14.
1q14.
3,12q12,16q13,19p,19q,22q2UNK+1p21.
1p21.
2,+1q31.
3p32.
1,2q37.
1,+3q25.
33,+4p16.
2p16.
3,+4p12p15.
31,+4q,+6p21.
2,+9p23,+12p11.
21p12.
1,+12q23,+12q24.
31,12q12,+16q23.
2q24.
2,+17q11.
2,18p,18q,19p,19q,+22q,+Xp,+Xq12332q37.
2,+3q21.
32p21.
33,+3p21.
31,+3q25.
33,+4p,+4q12,+4q35.
2,5p,5q,+6p22.
3,6q21,7p,7q11.
21q21.
2,7q33q35,+8p23.
2p23.
3,+8p12,+10p11.
21p15.
3,+10q11.
21q21.
1,+10q23.
1p26.
2,+12q21.
33q24.
33,+13q31.
1,+13q34,14q32.
33,15q11.
2q13.
1,+16q24.
2,16p12.
3p13.
2,16q22.
1q24.
2,17p,17q11.
2q12.
31,18p,18q,+19q13.
12,+20q11.
2111.
25,+20q13.
32q13.
33,22q4UNK+1q44,1q32.
1,+2p25.
3,+3q26.
2,3q26.
2,4q35.
2,+5q34,5q31.
2,+6p12.
3,8q12.
1,9p13.
1,9q12q13,+11p13,11p14.
1,11q21,+12q23.
1,+14q23.
3,14q11.
2,15q11.
2q25.
3,+16p13.
3,+16p11.
2,16p13.
3,16p12.
1,+21q21.
1q21.
352+1p35.
2p35.
3,+1p36.
11,1p36.
11,+3p22.
3,+3q26.
2,3p21.
31,3q26.
2,+4q27,+9p24.
3,+11p15.
5,+11q13.
1,+12q24.
31,12q14.
3,18q12.
2q12.
3,+13q34,+19p13.
263+1p34.
1p36.
33,+1p31.
2p32.
1,+1p21.
2p22.
3,+1q21.
1q21.
3,+1q22q24,+2q14.
1,+2q21.
1,2q31.
1,+3p14.
1p26.
3,+3q13.
11q29,+4q22.
1toq21.
22,5p11p15.
1,5q,6q12.
3,6q16.
3,6q21q22.
1,+7q22.
1,+8p,+8q22.
1q24.
3,+9p21.
1,+9p15.
2p13.
3,10q21.
3,11p15.
4,11p11.
12p11.
2,11q11q12.
2,11q13,q23–2,+12p13.
31p13.
33,+12q12q13.
12,+12q13.
13q14.
1,+12q14.
2q23.
1,+12q23.
3q24.
33,+13q14.
2q14.
3,14q12q23.
3,14q31.
1q31.
3,16p11.
2,16q11.
2,+17q21.
2,+17q24.
2,+17q25.
1,17q24.
3,18q12.
2q23,19p12,19q12,20p,20q,22q11.
21,Xp,Xq731q31.
2,6p,6q,7q31.
1,10q11.
21,+12p13.
31,12q24.
23q24.
31,12q24.
31,+13q,+14q24.
3,15q,16q12.
1q21,16q24.
2,+17p,+17q,19q13.
32,+21q,22q11.
23,Xp,Xq821p36.
22p36.
33,4p,4q,5p15.
33,+6p22.
1,+6p12.
3,6p22.
1,7p12.
3p13,+9,+9q21.
11q32.
2,+9q33.
3,+10p,+10q11.
21q23.
1,+10q23.
3210q26.
3,+11p13,12q24.
31,13q,14q12,21q,22q12.
11,+Xp21.
2p22.
33,+Xp11.
22p11.
4,+Xq93+1p36.
33,+1p36.
31,+1p33,1q23.
3,1q25.
2,2p11.
2,2q11.
2,2q22.
1q35,4p13p14,4q34.
1,+5p15.
2p15.
33,+5q35.
1q35.
3,6p,6q,+7p,+7q11.
2q21.
2,7q21.
3q36.
3,+9p13.
1,+9q12q13,+12p13.
31p13.
33,+12q24.
33,12q23.
3,+13q,+14q12,+15q13.
3,15q14q24.
1,15q14q26.
3,16p,16q,+18p,+18q,+19p,+19q,+20p,+20q,+21q,+22q,Xp12.
11231q23.
3,2p,2q,3q13.
2,+5p,+5q,+6p,+6q,+7q36.
1,+7q36.
3,9p13.
1,9q13,10p,10q11.
21q21.
3,+11q23.
3,12p13.
33,+13q,14q13.
2,14q24.
3,+15q11.
2q22.
31,+15q22.
32q26.
33,+16q12.
1q21,17p,17q,18q25,+19p13.
2,+19q12q13.
11,+20p,+20q,21q,Xp,Xq133+1q21.
2,2q33.
3,+5q31.
2q35.
3,+7q11.
23,+7q32.
1q32.
2,7p21.
3,+9p13.
3,9q21.
33,10p12.
1p12.
2,+11q12.
2,+11q23.
1q23.
2,11q23.
2q23.
3,+14q11.
2,17q21.
33q22,+19q13.
33,+19q13.
43,+20p,+20q,+Xp,+20q14UNK+1p36.
11,4q21.
21,+2p22.
3p23.
1,+2q24.
1,12q13.
2,12q14.
3q15,+16q12.
1,+19p13.
2,+22q12.
11521p35.
2p35.
3,1p11.
2p12,1q21.
1,1q31.
3q32.
1,4p12p13,6p21.
33,+7q11.
22q21.
11,9q31.
3,13q21.
2,17p13.
1,+21q22.
12q22.
3,+22q,+Xp,+XqMicroarrayanalysisofolfactoryneuroblastomaMGuledetal773ModernPathology(2008)21,770–778alterationsthatweresharedinatleasthalfofthesecaseswererecorded(Figure3).
LossofXp21.
1andgainsof20q11.
21–q11.
23,13q14.
2–q14.
3,and13q31.
1werepresentintwo-thirdsofstage3cancers.
Gainsat5q35and13q,andlossesat6q16–q22,2q31.
1,and2q33.
3,werepresentinhalfofthecases.
Thelongarmsofchromosomes13and20weregainedin50%oftheseolfactoryneuro-blastomas,possiblyindicatingimportantrolesintumorprogression.
DiscussionTheaimofourstudywastoidentifyDNAcopynumberchangesinolfactoryneuroblastomabyaCGH.
Untilrecently,genome-wide-profilingstu-dieshavereliedonconventionalCGHinchromo-somebandresolution.
Inonestudy,FISHandsingle-nucleotidepolymorphismmicroarrayanaly-siswereusedforonecaseofolfactoryneuroblasto-ma.
3Moreover,theCGHstudiesofcytogeneticaberrationsinolfactoryneuroblastomaarefew.
4–6Weperformedanoligonucleotide-basedaCGHana-lysison13olfactoryneuroblastomasamples,which,tothebestofourknowledge,wasthefirsttimethatarray-basedCGHwasappliedtostudythecopynumberchangesinthisneoplasm.
Severalcopynumberchangesreportedinpreviousstudieswereobservedinourstudy,withidentical,overlapping,orslightlydifferentminimalcommonregionsofalteration.
Gainsatthedistalpartsof1p,4,9p,13q,15q,22q,and21q,anddeletionsat4pandX,werereportedinatleastonestudy.
Gainsat7q11and20qanddeletionsat2q,5q,6p,6q,and18qweredetectedintwostudies(Tables3and4).
Overall,olfactoryneuroblastomashavehighlycomplexcopynumberchangesthatoccurovertheentiregenome.
Allsamplesanalyzedshowedgeno-micimbalanceswithslightlymoregainsthanlosses.
Asexpected,aCGHrevealedmorecopynumberchangesthanpreviousstudiesthatusedconven-tionalCGH.
Furthermore,ourresultsshowednovelaberrations,whichwerenotdescribedinpreviousTable3DNAcopynumbergainsinatleast20%ofthecasesdetectedbyaCGHChromosomelocationStart(kb)Stop(kb)Sizeofarea(kb)No.
ofgenesintheareaSuggestedtargetgenesSzymasetal(1997)Riazimandetal(2002)Bockmu¨hletal(2004)Hollandetal(2007)1p36.
316220638516541p32-pter1p35.
3281962835415831p35.
328625286896414p16.
2–p16.
31611661169444p12–p15.
311992748749288221324q1252152561554003274q21.
22–q22.
182376903828006714q27–q35.
2119605188460688553494q27–q35.
2a123408124209801385q3416087016169482465q35.
1–q35.
316787218070812836716p12.
34864514461446127q11.
23724407320976916LIMK1,FZD97q11.
27q11.
21,7q11.
237q21.
117699179296230569p13.
33506235844782319p13.
310p12.
3118796204301634712q23.
1948289745926311812q24.
311207381216338951813qWholeqarm13q14.
2–14.
3a478395175239135413q12.
11,13q33.
313q31.
1a78642800921450513q34a1129251132883638TFDP1,CUL4A13q3415q13.
32943930347908615q-qter16q12.
148851497859341116q20p/qWholechr.
20p/q20p12.
3–p12.
2a724186811440620q11.
1–q1220q11.
21–q11.
23a3041736618620112620q1320q13.
3120q13.
32–q13.
33a56224623006076110BRK21q22.
321qWholeqarm22q12.
12664327535892622qXp/qWholechr.
ThestartandendpointsoftheparticularaberrationwereestimatedfromCGHExplorersoftware.
NumberofgenesintheareaisaccordingtoNCBIdatabase.
Previouslyreportedresultsthatmatchthoseobtainedinthisstudyarealsoindicated.
aGainsinatleast30%ofthecases.
MicroarrayanalysisofolfactoryneuroblastomaMGuledetal774ModernPathology(2008)21,770–778reports.
Inaccordancewithatleasttwopreviousstudies,wefoundgainsat7q11.
2and20q13,andlossesat2q31–q37,5q,6p,6q,and18q.
Inadditiontothesepreviouslyreportedalterations,weidenti-fiednovelgainsinoursamplesat5q34–q35,6p12.
3,10p12.
31,12q23.
1–q24.
31,andallofchromosomeTable4DNAcopynumberlossesinatleast20%ofthecasesdetectedbyaCGHChromosomelocationStart(kb)Stop(kb)Sizeofarea(kb)No.
ofgenesintheareaSuggestedtargetgenesSzymasetal(1997)Riazimandetal(2002)Bockmu¨hletal(2004)Hollandetal(2007)2q31.
11734921752451753192q22–q322q31–q332q33.
3206359207454109518ADAM232q37.
12317752335281753382q37,2q37.
34p1341143445463403194p13–p15,4p/q5q31.
2138286138835549115q5q6p22.
126803282491446506p-pter6p21.
3330853318651012576p216p12.
347054509593905276q16.
3101732105348361626q14–q236q21107952110411245935FOXO3,CCNC6q22.
11134491166313182136q22–q2415q11.
2–q24.
1Wholeqarm15q13.
1a2762527834209218q12.
2–q12.
33199976014440151971818q18q12.
2–q12.
3a33268410407772919q1232846347761930619q13.
1141195414842891119q13.
3253405537913861019q13.
4363057635394822022q11.
2321670224938232122q12.
12650626849343322q11.
1–q11.
211539317451205848Xp/qWholechr.
XThestartandendpointsoftheparticularaberrationwereestimatedfromCGHExplorersoftware.
NumberofgenesintheareaisaccordingtoNCBIdatabase.
Previouslyreportedresultsthatmatchtheresultsobtainedbythisstudyarealsoindicated.
aLossesinatleast30%ofthecases.
12345678910111213141516171819202122XY0255075100255075100GenomicpositionAberrationfrequency-2q31.
1-2q33.
3-6q16.
3-q22.
1-6q12.
3-X-Xp21.
1+5q31.
3-q35.
3+13q14.
2-q14.
3+13q31.
1+13q+20q13.
32-q13.
33+20q11.
21-q11.
23+20GainsLossesFigure3Frequencyprofileofcopynumberaberrationsinsixstage3olfactoryneuroblastomasamples.
ThePiecewiseConstantFitalgorithmoftheCGHExplorersoftwarewasappliedtoidentifyDNAcopynumberalterations.
Frequenciesofcopynumbergains(gray)andlosses(black)areindicatedontheyaxis.
GenomicpositionsoftheaCGHprobesareindicatedonthexaxis.
Alterationsoccurringinatleast50%ofstage3casesaremarkedinthefigure.
MicroarrayanalysisofolfactoryneuroblastomaMGuledetal775ModernPathology(2008)21,770–778X.
Lossesat15q11.
2–q24.
1,15q13.
1,19q12–q13,22q11.
1–q11.
21,22q11.
23,and22q12.
1havenotbeendescribedpreviously.
Weidentifieda770kbregionofchromosomalgainat7q11.
2.
Thisregionhasbeenimplicatedinothercancers,andisoverexpressedinprostatecarcino-mas,adenoidcysticcarcinomas,headandnecksquamouscellcarcinomas,andpancreaticendo-crinetumors.
9–12CandidategeneslocatedwithinthisregionincludeLIMK1(NCBIGeneID:3984),apossibleoncogenethatcontributestocellcyclingandinvasion.
AnotherpossiblecandidateisFZD9(GeneID:8326),amemberofthe'frizzled'genefamilythatisupregulatedinastrocytomasandgastriccarcinomas.
13,14A6Mbregionofgainat20q13.
32–q13.
33wasalsoidentified.
DNAcopynumberincreasesatchromo-some20q13havebeenobservedfrequentlyinavarietyofcancers,includingbreast,ovarian,andsquamouscellcarcinomas,15suggestingthattheregionharborsoneormoreoncogenes.
Inparticular,20q13.
2hasbeenproposedasahotspotforcandi-dategenes.
15Itshouldalsobenotedthatover-expressionatthislocushasbeenassociatedwithreducedpatientsurvivalandhighertumorgrade.
20q13.
3,frequentlyoverexpressedinovariancan-cers,hasbeenrecentlyshowntolocalizetheBRKtyrosinekinasegene(GeneID:5753),whichisthoughttohaveanimportantroleinthedevelop-mentofovariancancers.
16Lossesoccurringatchromosome2qhavebeendescribedforvariouscarcinomas,includingheadandnecksquamouscellcarcinoma,17–19breastcarcinoma,20lungcarcinoma,21neuroblastoma,22cervicalcancer,23andprostateadenocarcinoma.
24Studiesusingdifferentapproacheshaveincreas-inglyshownthatthemostaffectedregionis2q32–q37.
Thisregionalsoseemstobeimpli-catedinthedevelopmentofONB,asithasbeenreportedinthreecytogeneticstudiesincludingourinvestigation.
Severalcandidatetumorsuppressorgeneshavebeensuggested,includingADAM23(GeneID:8745),thoughttofunctionasanadhesionmolecule,whichpromotestheattachmentofneuralcells.
25Thisgeneat2q33.
3waslostin20%ofallolfactoryneuroblastomasamplesanalyzedand,moresignificantly,in50%ofstage3tumors.
Takadaetalhavesuggestedanessentialroleforthiscandidatetumorsuppressorgeneintheprogressionofgastriccancer,andourresultsindicatethatthismightbethecaseforolfactoryneuroblastomaaswell.
Anotherareaoflossidentifiedinourstudy,andalsoreportedbyBockmu¨hletal4andHollandetal,3islocatedat6q21–22.
Thisregionisfrequentlydeletedinavarietyofneoplasms,includingpan-creaticendocrinetumors,26prostatecarcinoma,27breastcarcinoma,28andcentralnervoussystemlymphomas.
29Threemainputativetumorsuppres-sorgeneshavebeenproposed,includingFOXO3(GeneID:2309),CCNC(GeneID:892),andPTPRK(GeneID:5796).
Thefirsttwocandidategenesareintheregionsfoundtobedeletedinoursamples.
Ourstudyalsoidentifiedtwosmallgainsat9p13.
3(782kb)and13q34(363kb)thatwerepreviouslyreportedbyHollandetal.
The9p13.
3locushasbeenshowntobegainedinprostatecancercelllinesintworecentstudiesusingaCGH.
30,31Theseauthorssuggestedtheinvolvementofthreegenes,includingUBE2R2(GeneID:54926),DCTN3(GeneID:11258),andIL-11RA(GeneID:3590).
Kamradtetalscreened20primaryprostatecancersamplesandfoundthatonlyIL-11RAwasgainedin75%ofthetumors,whereasonlyDCTN3wasnotgainedinanyofthecases;bothgenesweregainedtogetherin10%ofthetumors.
ThisobservationledtheauthorstosuggestthatIL-11RAisthetargetofgainratherthanDCTN3.
Gainsat13q34havealsobeendescribedpreviouslyindifferentcancers,includingbreastcancer,hepatocellularcarcinoma,esophagealsquamouscellcarcinoma,andlungadenocarcinoma.
32TheTFDP1(GeneID:7027)andCUL4A(GeneID:8451)genesweresuggestedastargetgenesthatpossiblyhavearoleincarcinogen-esis.
Furtherstudiesarerequiredtodetermineifthesegenesareinvolvedinolfactoryneuroblastoma.
Asformosttumors,stageisthemostimportantparameterassociatedwithsurvivalinolfactoryneuroblastoma.
2Ourresultsclearlyindicatethatalterationsin20qand13qareimportantintheprogressionofolfactoryneuroblastoma.
Gainof20qhasbeenwidelyassociatedwithprogressionofseveraltumors,includingbreastcarcinoma,33cervi-calcarcinoma,34andpancreaticcarcinoma.
35Bothlossesandgainsofchromosome13qhavebeennotedinmanyrecentstudiesofvarioustumors,suggestingtheexistenceofnoveloncogenesortumorsuppressorgenesorbothinthisregion.
Furthermore,thisregionhasbeenreportedtocontainmicroRNAsthatcouldfunctionastumorsuppressorgenesoroncogenes.
36Theonlyknowntumor-relatedgeneinthisregionisthetumorsuppressorgeneRB1.
Itisassociatedwithlossoffunctionordecreasedgeneexpressionintumorcellsand,consequently,cannotbeconsideredfurther.
Lillingtonetal37foundthatgainof13qismorefrequentinolderpatients,whosetumorsdisplaymoreabnormalitiesthanthoseinyoungerpatients.
Theseauthorshavealsoreportedthatgainof13qisseenmorefrequentlyinpoorlydifferentiatedmalig-nancies,whichtypicallycontainmoreabnormalitiesthanwell-differentiatedcancers.
Gainsofboth13qand20qareseenincolorectalcarcinomasandtheirprogression.
38,39Inconclusion,wehaveidentifiednovelchromo-somalregions,whicharefrequentlyalteredinolfactoryneuroblastoma.
Whentheresultsofourstudyarecombinedwithpreviouslypublisheddata,severalregionsareidentifiedthatareconsistentlyabnormalinolfactoryneuroblastoma.
Theseregionsarealsofrequentlyassociatedwithtumorigenesisofavarietyofcancers,especiallycarcinomas.
AMicroarrayanalysisofolfactoryneuroblastomaMGuledetal776ModernPathology(2008)21,770–778numberofgeneslocatedatthesesiteshavebeensuggestedtobeimportant,butfurtherexperimentsarerequiredtodeterminetheirrolesinolfactoryneuroblastoma.
DisclosureTheauthorsstatenoconflictofinterest.
References1BarnesL,(ed)PathologyandGeneticsofHeadandNeckTumours.
InternationalAgencyforResearchonCancer(IARC),Lyon,France,2005,pp430.
2MoritaA,EbersoldMJ,OlsenKD,etal.
Esthesioneur-oblastoma:prognosisandmanagement.
Neurosurgery1993;32:706–714,discussion714–715.
3HollandH,KoschnyR,KruppW,etal.
Comprehensivecytogeneticcharacterizationofanesthesioneuroblas-toma.
CancerGenetCytogenet2007;173:89–96.
4BockmuhlU,YouX,Pacyna-GengelbachM,etal.
CGHpatternofesthesioneuroblastomaandtheirmetastases.
BrainPathol2004;14:158–163.
5RiazimandSH,BriegerJ,JacobR,etal.
Analysisofcytogeneticaberrationsinesthesioneuroblastomasbycomparativegenomichybridization.
CancerGenetCytogenet2002;136:53–57.
6SzymasJ,WolfG,KowalczykD,etal.
Olfactoryneuroblastoma:detectionofgenomicimbalancesbycomparativegenomichybridization.
ActaNeurochir(Wien)1997;139:839–844.
7MillsSE,FriersonJrHF.
Olfactoryneuroblastoma.
Aclinicopathologicstudyof21cases.
AmJSurgPathol1985;9:317–327.
8LingjaerdeOC,BaumbuschLO,LiestolK,etal.
CGH-explorer:aprogramforanalysisofarray-CGHdata.
Bioinformatics2005;21:821–822.
9DavilaM,FrostAR,GrizzleWE,etal.
LIMkinase1isessentialfortheinvasivegrowthofprostateepithelialcells:implicationsinprostatecancer.
JBiolChem2003;278:36868–36875.
10KasamatsuA,EndoY,UzawaK,etal.
Identificationofcandidategenesassociatedwithsalivaryadenoidcysticcarcinomasusingcombinedcomparativegeno-michybridizationandoligonucleotidemicroarrayanalyses.
IntJBiochemCellBiol2005;37:1869–1880.
11BockmuhlU,SchlunsK,SchmidtS,etal.
Chromo-somalalterationsduringmetastasisformationofheadandnecksquamouscellcarcinoma.
GenesChromo-somesCancer2002;33:29–35.
12StumpfE,AaltoY,HoogA,etal.
Chromosomalalterationsinhumanpancreaticendocrinetumors.
GenesChromosomesCancer2000;29:83–87.
13ZhangZ,SchittenhelmJ,GuoK,etal.
Upregulationoffrizzled9inastrocytomas.
NeuropatholApplNeuro-biol2006;32:615–624.
14KirikoshiH,SekiharaH,KatohM.
Expressionprofilesof10membersoffrizzledgenefamilyinhumangastriccancer.
IntJOncol2001;19:767–771.
15QuinlanKG,VergerA,YaswenP,etal.
Amplificationofzincfingergene217(ZNF217)andcancer:whengoodfingersgobad.
BiochimBiophysActa2007;1775:333–340.
16SchmandtRE,BennettM,CliffordS,etal.
TheBRKtyrosinekinaseisexpressedinhigh-gradeserouscarcinomaoftheovary.
CancerBiolTher2006;5:1136–1141.
17BederLB,GunduzM,OuchidaM,etal.
Genome-wideanalysesonlossofheterozygosityinheadandnecksquamouscellcarcinomas.
LabInvest2003;83:99–105.
18YamamotoN,MizoeJ,NumasawaH,etal.
Alleliclossonchromosomes2q,3pand21q:possiblyapoorprognosticfactorinoralsquamouscellcarcinoma.
OralOncol2003;39:796–805.
19CengizB,GunduzM,NagatsukaH,etal.
Finedeletionmappingofchromosome2q21–37showsthreeprefer-entiallydeletedregionsinoralcancer.
OralOncol2007;43:241–247.
20PiaoZ,LeeKS,KimH,etal.
Identificationofnoveldeletionregionsonchromosomearms2qand6pinbreastcarcinomasbyamplotypeanalysis.
GenesChromosomesCancer2001;30:113–122.
21OtsukaT,KohnoT,MoriM,etal.
Deletionmappingofchromosome2inhumanlungcarcinoma.
GenesChromosomesCancer1996;16:113–119.
22TakitaJ,YangHW,ChenYY,etal.
Allelicimbalanceonchromosome2qandalterationsofthecaspase8geneinneuroblastoma.
Oncogene2001;20:4424–4432.
23NarayanG,PulidoHA,KoulS,etal.
Geneticanalysisidentifiesputativetumorsuppressorsitesat2q35–q36.
1and2q36.
3–q37.
1involvedincervicalcancerprogression.
Oncogene2003;22:3489–3499.
24UedaT,KomiyaA,SuzukiH,etal.
Lossofhetero-zygosityonchromosome2injapanesepatientswithprostatecancer.
Prostate2005;64:265–271.
25TakadaH,ImotoI,TsudaH,etal.
ADAM23,apossibletumorsuppressorgene,isfrequentlysilencedingastriccancersbyhomozygousdeletionoraberrantpromoterhypermethylation.
Oncogene2005;24:8051–8060.
26BarghornA,SpeelEJ,FarspourB,etal.
Putativetumorsuppressorlociat6q22and6q23–q24areinvolvedinthemalignantprogressionofsporadicendocrinepancreatictumors.
AmJPathol2001;158:1903–1911.
27KimJH,DhanasekaranSM,MehraR,etal.
Integrativeanalysisofgenomicaberrationsassociatedwithpros-tatecancerprogression.
CancerRes2007;67:8229–8239.
28TheileM,SeitzS,ArnoldW,etal.
Adefinedchromosome6qfragment(atD6S310)harborsaputativetumorsuppressorgeneforbreastcancer.
Oncogene1996;13:677–685.
29NakamuraM,KishiM,SakakiT,etal.
Noveltumorsuppressorlocion6q22–23inprimarycentralnervoussystemlymphomas.
CancerRes2003;63:737–741.
30SaramakiOR,PorkkaKP,VessellaRL,etal.
Geneticaberrationsinprostatecancerbymicroarrayanalysis.
IntJCancer2006;119:1322–1329.
31KamradtJ,JungV,WahrheitK,etal.
Detectionofnovelampliconsinprostatecancerbycomprehensivegenomicprofilingofprostatecancercelllinesusingoligonucleotide-basedarrayCGH.
PLoSONE2007;2:e769.
3232AbbaMC,FabrisVT,HuY,etal.
Identificationofnovelamplificationgenetargetsinmouseandhumanbreastcanceratasyntenicclustermappingtomousech8A1andhumanch13q34.
CancerRes2007;67:4104–4112.
33HodgsonJG,ChinK,CollinsC,etal.
Genomeamplificationofchromosome20inbreastcancer.
BreastCancerResTreat2003;78:337–345.
MicroarrayanalysisofolfactoryneuroblastomaMGuledetal777ModernPathology(2008)21,770–77834WiltingSM,SnijdersPJ,MeijerGA,etal.
Increasedgenecopynumbersatchromosome20qarefrequentinbothsquamouscellcarcinomasandadenocarcinomasofthecervix.
JPathol2006;209:220–230.
35LoukopoulosP,ShibataT,KatohH,etal.
Genome-widearray-basedcomparativegenomichybridizationanalysisofpancreaticadenocarcinoma:identificationofgeneticindicatorsthatpredictpatientoutcome.
CancerSci2007;98:392–400.
36CalinGA,CroceCM.
MicroRNAsandchromosomalabnormalitiesincancercells.
Oncogene2006;25:6202–6210.
37LillingtonDM,KingstonJE,CoenPG,etal.
Compara-tivegenomichybridizationof49primaryretinoblasto-matumorsidentifieschromosomalregionsassociatedwithhistopathology,progression,andpatientout-come.
GenesChromosomesCancer2003;36:121–128.
38LipsEH,deGraafEJ,TollenaarRA,etal.
Singlenucleotidepolymorphismarrayanalysisofchromoso-malinstabilitypatternsdiscriminatesrectaladenomasfromcarcinomas.
JPathol2007;212:269–277.
39HermsenM,PostmaC,BaakJ,etal.
Colorectaladenomatocarcinomaprogressionfollowsmultiplepathwaysofchromosomalinstability.
Gastroenterology2002;123:1109–1119.
MicroarrayanalysisofolfactoryneuroblastomaMGuledetal778ModernPathology(2008)21,770–778
Duetoitsrarity,littleisunderstoodregardingitsmolecularandcytogeneticabnormalities.
TheaimofthecurrentstudyistoidentifyspecificDNAcopynumberchangesinolfactoryneuroblastoma.
Thirteendissectedtissuesampleswereanalyzedusingarraycomparativegenomichybridization.
Ourresultsshowthatgenecopynumberprofilesofolfactoryneuroblastomasamplesarecomplex.
Themostfrequentchangesincludedgainsat7q11.
22–q21.
11,9p13.
3,13q,20p/q,andXp/q,andlossesat2q31.
1,2q33.
3,2q37.
1,6q16.
3,6q21.
33,6q22.
1,22q11.
23,22q12.
1,andXp/q.
Gainsweremorefrequentthanlosses,andhigh-stagetumorsshowedmorealterationsthanlow-stageolfactoryneuroblastoma.
Frequentchangesinhigh-stagetumorsweregainsat13q14.
2–q14.
3,13q31.
1,and20q11.
21–q11.
23,andlossofXp21.
1(in66%ofcases).
Gainsat5q35,13q,and20q,andlossesat2q31.
1,2q33.
3,and6q16–q22,werepresentin50%ofcases.
Theidentifiedregionsofgenecopynumberchangehavebeenimplicatedinavarietyoftumors,especiallycarcinomas.
Inaddition,ourresultsindicatethatgainsin20qand13qmaybeimportantintheprogressionofthiscancer,andthattheseregionspossiblyharborgeneswithfunctionalrelevanceinolfactoryneuroblastoma.
ModernPathology(2008)21,770–778;doi:10.
1038/modpathol.
2008.
57;publishedonline11April2008Keywords:olfactoryneuroblastoma;arrayCGH;esthesioneuroblastoma;cytogeneticsOlfactoryneuroblastomaisrare,withanestimatedincidenceof0.
4permillionpeopleperyear.
Itaccountsforapproximately2–3%ofsinonasaltracttumors.
1Olfactoryneuroblastomaisamalignantneuroectodermaltumor,whichisbelievedtoorigi-nateattheolfactorymembraneofthesinonasaltract.
1Symptomsincludeunilateralnasalobstruc-tionandepistaxis.
TheKadishsystemclassifiescasesofolfactoryneuroblastomaintothreestages.
Stage1consistsoftumorsconfinedtothenasalcavity;stage2tumorsextendtotheparanasalsinuses;andstage3neoplasmsextendbeyondthesinonasalcavities(somehaveaddedastage4forthosetumorswithmetastases).
Patientswithhigh-stagetumorshaveaworseprognosiscomparedwiththosewithlow-stagetumors,as5-yearsurvivalratesare40and80%,respectively.
2Cytogeneticdataforolfactoryneuroblastomaarelimited.
ThemostrecentlypublishedstudywascarriedoutbyHollandetal,3whoperformedcytogeneticcharacterizationofonecaseusingtrypsinGiemsastaining(GTGbanding),multicolorfluorescenceinsituhybridization(M-FISH),andsingle-nucleotidepolymorphismkaryotyping.
Theyreportednumerouschromosomalaberrationspre-dominantlyinvolvingchromosomes2q,5,6q,17,19,21q,and22,aswellastrisomy8.
Bockmu¨ehletal4appliedconventionalcomparativegenomichy-bridization(CGH)to22olfactoryneuroblastomasandreportedfrequentdeletionsof1p,3p/q,9p,and10p/q,andamplificationsof17q,17p13,20p,and22q.
Theyalsonotedaspecificdeletiononchromo-some11andgainonchromosome1p,whichwereassociatedwithmetastasisandaworseprognosis.
ThreeolfactoryneuroblastomaswerestudiedbyRiazimandetal5usingconventionalCGH,andamplificationofwholechromosome19,partialgainsof1p,8q,15q,and22q,anddeletionsof4qand6pReceived24November2007;revised02March2008;accepted10March2008;publishedonline11April2008Correspondence:DrEBStelow,MD,DepartmentofPathology,UniversityofVirginiaHealthSciences,Box800214,JeffersonParkAvenue,Charlottesville,VA22908,USA.
E-mail:edstelow@yahoo.
comandDrSKnuutila,PhD,DepartmentofPathology,HaartmanInstituteandHUSLAB,POBox21(Haartmaninkatu3),UniversityofHelsinki,FI-00014Helsinki,Finland.
E-mail:Sakari.
Knuutila@helsinki.
fiModernPathology(2008)21,770–778&2008USCAP,IncAllrightsreserved0893-3952/08$30.
00www.
modernpathology.
orgweredetected.
Szymasetal6studiedasingleolfactoryneuroblastomaandfoundgainsofwholechromosomes4,8,11,and14,partialgainsof1qand17q,partialdeletionsof5qand17q,andwholechromosomelossesof16,18,19,andX.
Inourstudy,weappliedforthefirsttimeanoligonucleo-tide-basedarrayCGH(aCGH)toidentifythemostfrequentlyoccurringDNAcopynumberchangesin13casesofolfactoryneuroblastoma.
Weidentifiednovelchromosomalregionsthatwerefrequentlyalteredinadditiontopreviouslyreportedabnormalregions.
MaterialsandmethodsTumorSamplesTheSurgicalPathologyDatabase(DepartmentofPathologyattheUniversityofVirginia,Charlottes-ville,VA,USA)wassearchedforresectedcasesofolfactoryneuroblastoma.
Slidesfromformalin-fixed,paraffin-embeddedtissueswerereviewed,and15caseswereselectedbasedontheavailabilityofabundantandwell-preservedneoplastictissue.
Onlycaseswithconventionalhistology,asde-scribedbyMillsandFrierson,7werechosen(Figure1).
Allcasesexhibitedatypicalimmunophenotypeandwereimmunoreactivewithantibodiestosynap-tophysinandnotwithantibodiestokeratins.
Areasrepresentingcancertissueweredissectedfromparaffin-embeddedtumorsusingconventionallightmicroscopyandscalpelblades.
All15samplessubmittedforanalysiswerecomposedofatleast50%tumorcells.
EachofsixsampleswasclassifiedasKadishstage2and3(Figure1),whereasinformationregardingthestagewasnotavailableforthreetumors.
Patients'agerangedfrom27to62years.
TheclinicopathologicaldataforthepatientsandsamplesaresummarizedinTable1.
DNAExtractionandDigestionTotalDNAwasextractedusingQIAampDNAminikit,accordingtothemanufacturer'sinstructions(Qiagen,Hilden,Germany).
DNAconcentrationsoftheextractedsamplesweredeterminedusingtheNanoDropND-1000spectrophotometer(Nanodroptechnologies,Wilmington,DE,USA)at260/280nm.
TheintegrityoftheextractedDNAwasverifiedbyagarosegelelectrophoresis(datanotshown).
A1.
5mgquantityofgenomicDNAfromtestandreferencesampleswasdigestedusingAlu1andRsa1restrictionenzymes(Promega,Madison,WI,USA)for2hat371C.
Toinactivatetheenzymes,thesampleswereincubatedat651Cfor20min.
ArrayCGHExperimentsArrayCGHwasperformedfor13samplesthathadgoodqualityDNAwithsufficientyields.
ForFigure1StandardH&Esectionsofconventionalolfactoryneuroblastoma:(a)sample11(100),(b)sample12(200).
Table1ClinicopathologicaldataforthesamplesSampleSex/ageDead/aliveDiseasestatusTime(months)Stage(Kadish)1m/50DWD15622m/49ANED192UNK3m/53DWD2034f/38AUNK136UNK5f/40AUNK14926f/27ANED11937f/61ANED10638m/27ANED5929m/40ANED57312f/62DWD35313m/41UNKUNKUNK314m/44ANED24UNK15m/60ANED62Abbreviations:A,alive;D,dead;NED,noevidenceofdisease;UNK,unknown;WD,withdisease.
MicroarrayanalysisofolfactoryneuroblastomaMGuledetal771ModernPathology(2008)21,770–778reference,pooledDNAextractedfrombuffycoatfractionsofwholebloodobtainedfromsex-matched,healthyindividualsprovidedbytheFinnishRedCrosswasused.
TheAgilentgenomicDNAlabelingkitPLUS(AgilentTechnologies,SantaClara,CA,USA)wasusedtolabel1.
5mgofdigestedgenomicDNA.
TheDNAofolfactoryneuroblastomasamplesandthereferenceDNAwerelabeledwithcyanine5-dUTPandcyanine3-dUTPfluorochromes,respec-tively,for2hat371C.
Forinactivationoftheenzymes,thesampleswereincubatedat651Cfor10min.
Humancot-1DNA(Invitrogen,Carlsbad,CA,USA)wasaddedtothereactionsfor3minat951Candthenfor30minat371C.
Priortohybridiza-tionexperiments,labeledDNAwascleanedaccord-ingtoAgilent'sprotocol.
LabeledsampleandreferenceDNAwerehybridizedonAgilent's444KCGHmicroarray(productnumberG4426AandG4426B,AgilentTechnologies)for24hat651C.
Priortoscanning,microarrayslideswerewashedaccordingtothemanufacturer'sinstructions.
ThemicroarrayslideswerescannedusingDNAMicro-arrayScannerandthescannedimageswereana-lyzedusingtheFeatureExtractionSoftware(AgilentTechnologies).
DatawereimportedtotheCGHAnalyticsSoftware3.
4foranalysisofindividualcases.
Z-scorealgorithmwiththresholdat3.
5wasusedintheidentificationofcopynumberchanges.
Toobtainaglobalviewofthecopynumberalterationfrequenciesinolfactoryneuroblastoma,wealsoanalyzedthedatausingGeneSpring(AgilentTechnologies)andCGHExplorersoftware3.
1.
8TherawdatafromtheFeatureExtractionsoftwarewereimportedintotheGeneSpringsoftwareandnormal-izedusingtheFeatureExtractiondataimportplug-in(AgilentTechnologies).
OutlierprobesmarkedbyFeatureExtractionwereignoredinnormalization.
Samplespotintensitiesweredividedbycontrolchannelintensities,andmicroarrayswerenormal-izedtothe50thpercentileandprobestothemedianvalue.
Outlierprobeswerefilteredfromfurtheranalysis.
Normalizedandfiltereddatawereim-portedintotheCGHExplorersoftware.
CopynumberaberrationswereidentifiedusingPiecewiseConstantFit(PCF)algorithm.
Defaultparameterswereappliedtodetectlongcopynumberaberra-tions.
Thesignificancethresholdwassetat0.
2.
ResultsGeneCopyNumberProfilingofOlfactoryNeuroblastomaArrayCGHwasappliedtostudygenecopynumberalterationsin13olfactoryneuroblastomasamples.
TheCGHExplorersoftwarewasusedtoanalyzeglobalfrequenciesofcopynumberchange(Figure2).
Atotalof15.
74%oftheprobeswerefoundaberrantwhenathresholdof0.
2wasusedinthePCFanalysis.
DNAcopynumbergains(8.
37%)wereslightlymorefrequentthanlosses(7.
36%).
Inaddition,theCGHanalyticssoftwarewasusedtodeterminecopynumberaberrationsinindividualsamples.
CopynumberaberrationsforeachcasearesummarizedinTable2.
Changesinthecopynumberofentirechromosomes(aneuploidy)orchromosome12345678910111213141516171819202122XY0255075100255075100AberrationfrequencyGenomicpositionGainsLossesFigure2Frequencyprofileofcopynumberaberrationsin13olfactoryneuroblastomasamples.
CGHExplorersoftwareandPiecewiseConstantFitalgorithmwereusedtodeterminecopynumberaberrationsinolfactoryneuroblastomasamples.
Thechromosomalalterationsareshownineachprobepositionasincidencebar.
Gainsofgenomicmaterialareindicatedingrayontheuppersideofthemiddleline(at0).
Lossesareindicatedinblackonthebottomsideofthemiddleline.
GenomicpositionsoftheaCGHprobesaremarkedonthexaxis.
MicroarrayanalysisofolfactoryneuroblastomaMGuledetal772ModernPathology(2008)21,770–778armsrepresented16%ofallalterations;interest-ingly,themostalteredwholechromosomeswere20,21,22,andX,andalmost70%oftheaberrationsofthesewerewholechromosomelossesorgains.
Toobtainamoredetailedoverviewofthemostfrequentlyoccurringalterations,onlythosealtera-tionssharedbyatleast20%ofthecaseswererecorded.
Thisapproachidentifiedanequalnumberofgainedandlostchromosomalregions(n21).
Tables3and4showthesizeofthealteredareasandwhethertheyhavebeenreportedinpreviouscopynumberchangestudiesforolfactoryneuroblastoma.
Thetablesalsoincludesuggestedtargetgenesthatarelocatedatthesitesofalterationandthatmayplayaroleintumorigenesis.
SpecificCopyNumberAlterationsWereIdentifiedinCasesofOlfactoryNeuroblastomaofDifferentStageAstage3olfactoryneuroblastomashowedthemostDNAcopynumberchanges,with59%ofallalterations.
Also,mostaneuploidyoccurredinhigh-stagetumors;70%ofwhole-chromosomeaber-rationswereidentifiedinthesetumors.
Forstage3tumors,themeanDNAcopynumberchangepercasewas28.
5,whereasstage2olfactoryneuroblas-tomashadameannumberof17changespertumor.
However,only4outof6stage2casescontainedsufficientgoodqualityDNAtoperformaCGH.
Tofurthercharacterizethegenomicchangesthatoccurredinalater-stageolfactoryneuroblastoma,Table2aCGHresultsfor13ONBsamplesONBsampleStageTumorDNAcopynumberchanges12+1p31.
2,+1p31.
1,+1p35.
3,+1q31.
1q31.
3,+2p24.
3,2p25.
1p25.
3,2q36.
3q37.
3,+3p13.
12p13.
31,+4p,+4q,+5q23.
1,+8q22.
3q24.
12,9p21.
1,10p12.
31,11q14.
1q14.
3,12q12,16q13,19p,19q,22q2UNK+1p21.
1p21.
2,+1q31.
3p32.
1,2q37.
1,+3q25.
33,+4p16.
2p16.
3,+4p12p15.
31,+4q,+6p21.
2,+9p23,+12p11.
21p12.
1,+12q23,+12q24.
31,12q12,+16q23.
2q24.
2,+17q11.
2,18p,18q,19p,19q,+22q,+Xp,+Xq12332q37.
2,+3q21.
32p21.
33,+3p21.
31,+3q25.
33,+4p,+4q12,+4q35.
2,5p,5q,+6p22.
3,6q21,7p,7q11.
21q21.
2,7q33q35,+8p23.
2p23.
3,+8p12,+10p11.
21p15.
3,+10q11.
21q21.
1,+10q23.
1p26.
2,+12q21.
33q24.
33,+13q31.
1,+13q34,14q32.
33,15q11.
2q13.
1,+16q24.
2,16p12.
3p13.
2,16q22.
1q24.
2,17p,17q11.
2q12.
31,18p,18q,+19q13.
12,+20q11.
2111.
25,+20q13.
32q13.
33,22q4UNK+1q44,1q32.
1,+2p25.
3,+3q26.
2,3q26.
2,4q35.
2,+5q34,5q31.
2,+6p12.
3,8q12.
1,9p13.
1,9q12q13,+11p13,11p14.
1,11q21,+12q23.
1,+14q23.
3,14q11.
2,15q11.
2q25.
3,+16p13.
3,+16p11.
2,16p13.
3,16p12.
1,+21q21.
1q21.
352+1p35.
2p35.
3,+1p36.
11,1p36.
11,+3p22.
3,+3q26.
2,3p21.
31,3q26.
2,+4q27,+9p24.
3,+11p15.
5,+11q13.
1,+12q24.
31,12q14.
3,18q12.
2q12.
3,+13q34,+19p13.
263+1p34.
1p36.
33,+1p31.
2p32.
1,+1p21.
2p22.
3,+1q21.
1q21.
3,+1q22q24,+2q14.
1,+2q21.
1,2q31.
1,+3p14.
1p26.
3,+3q13.
11q29,+4q22.
1toq21.
22,5p11p15.
1,5q,6q12.
3,6q16.
3,6q21q22.
1,+7q22.
1,+8p,+8q22.
1q24.
3,+9p21.
1,+9p15.
2p13.
3,10q21.
3,11p15.
4,11p11.
12p11.
2,11q11q12.
2,11q13,q23–2,+12p13.
31p13.
33,+12q12q13.
12,+12q13.
13q14.
1,+12q14.
2q23.
1,+12q23.
3q24.
33,+13q14.
2q14.
3,14q12q23.
3,14q31.
1q31.
3,16p11.
2,16q11.
2,+17q21.
2,+17q24.
2,+17q25.
1,17q24.
3,18q12.
2q23,19p12,19q12,20p,20q,22q11.
21,Xp,Xq731q31.
2,6p,6q,7q31.
1,10q11.
21,+12p13.
31,12q24.
23q24.
31,12q24.
31,+13q,+14q24.
3,15q,16q12.
1q21,16q24.
2,+17p,+17q,19q13.
32,+21q,22q11.
23,Xp,Xq821p36.
22p36.
33,4p,4q,5p15.
33,+6p22.
1,+6p12.
3,6p22.
1,7p12.
3p13,+9,+9q21.
11q32.
2,+9q33.
3,+10p,+10q11.
21q23.
1,+10q23.
3210q26.
3,+11p13,12q24.
31,13q,14q12,21q,22q12.
11,+Xp21.
2p22.
33,+Xp11.
22p11.
4,+Xq93+1p36.
33,+1p36.
31,+1p33,1q23.
3,1q25.
2,2p11.
2,2q11.
2,2q22.
1q35,4p13p14,4q34.
1,+5p15.
2p15.
33,+5q35.
1q35.
3,6p,6q,+7p,+7q11.
2q21.
2,7q21.
3q36.
3,+9p13.
1,+9q12q13,+12p13.
31p13.
33,+12q24.
33,12q23.
3,+13q,+14q12,+15q13.
3,15q14q24.
1,15q14q26.
3,16p,16q,+18p,+18q,+19p,+19q,+20p,+20q,+21q,+22q,Xp12.
11231q23.
3,2p,2q,3q13.
2,+5p,+5q,+6p,+6q,+7q36.
1,+7q36.
3,9p13.
1,9q13,10p,10q11.
21q21.
3,+11q23.
3,12p13.
33,+13q,14q13.
2,14q24.
3,+15q11.
2q22.
31,+15q22.
32q26.
33,+16q12.
1q21,17p,17q,18q25,+19p13.
2,+19q12q13.
11,+20p,+20q,21q,Xp,Xq133+1q21.
2,2q33.
3,+5q31.
2q35.
3,+7q11.
23,+7q32.
1q32.
2,7p21.
3,+9p13.
3,9q21.
33,10p12.
1p12.
2,+11q12.
2,+11q23.
1q23.
2,11q23.
2q23.
3,+14q11.
2,17q21.
33q22,+19q13.
33,+19q13.
43,+20p,+20q,+Xp,+20q14UNK+1p36.
11,4q21.
21,+2p22.
3p23.
1,+2q24.
1,12q13.
2,12q14.
3q15,+16q12.
1,+19p13.
2,+22q12.
11521p35.
2p35.
3,1p11.
2p12,1q21.
1,1q31.
3q32.
1,4p12p13,6p21.
33,+7q11.
22q21.
11,9q31.
3,13q21.
2,17p13.
1,+21q22.
12q22.
3,+22q,+Xp,+XqMicroarrayanalysisofolfactoryneuroblastomaMGuledetal773ModernPathology(2008)21,770–778alterationsthatweresharedinatleasthalfofthesecaseswererecorded(Figure3).
LossofXp21.
1andgainsof20q11.
21–q11.
23,13q14.
2–q14.
3,and13q31.
1werepresentintwo-thirdsofstage3cancers.
Gainsat5q35and13q,andlossesat6q16–q22,2q31.
1,and2q33.
3,werepresentinhalfofthecases.
Thelongarmsofchromosomes13and20weregainedin50%oftheseolfactoryneuro-blastomas,possiblyindicatingimportantrolesintumorprogression.
DiscussionTheaimofourstudywastoidentifyDNAcopynumberchangesinolfactoryneuroblastomabyaCGH.
Untilrecently,genome-wide-profilingstu-dieshavereliedonconventionalCGHinchromo-somebandresolution.
Inonestudy,FISHandsingle-nucleotidepolymorphismmicroarrayanaly-siswereusedforonecaseofolfactoryneuroblasto-ma.
3Moreover,theCGHstudiesofcytogeneticaberrationsinolfactoryneuroblastomaarefew.
4–6Weperformedanoligonucleotide-basedaCGHana-lysison13olfactoryneuroblastomasamples,which,tothebestofourknowledge,wasthefirsttimethatarray-basedCGHwasappliedtostudythecopynumberchangesinthisneoplasm.
Severalcopynumberchangesreportedinpreviousstudieswereobservedinourstudy,withidentical,overlapping,orslightlydifferentminimalcommonregionsofalteration.
Gainsatthedistalpartsof1p,4,9p,13q,15q,22q,and21q,anddeletionsat4pandX,werereportedinatleastonestudy.
Gainsat7q11and20qanddeletionsat2q,5q,6p,6q,and18qweredetectedintwostudies(Tables3and4).
Overall,olfactoryneuroblastomashavehighlycomplexcopynumberchangesthatoccurovertheentiregenome.
Allsamplesanalyzedshowedgeno-micimbalanceswithslightlymoregainsthanlosses.
Asexpected,aCGHrevealedmorecopynumberchangesthanpreviousstudiesthatusedconven-tionalCGH.
Furthermore,ourresultsshowednovelaberrations,whichwerenotdescribedinpreviousTable3DNAcopynumbergainsinatleast20%ofthecasesdetectedbyaCGHChromosomelocationStart(kb)Stop(kb)Sizeofarea(kb)No.
ofgenesintheareaSuggestedtargetgenesSzymasetal(1997)Riazimandetal(2002)Bockmu¨hletal(2004)Hollandetal(2007)1p36.
316220638516541p32-pter1p35.
3281962835415831p35.
328625286896414p16.
2–p16.
31611661169444p12–p15.
311992748749288221324q1252152561554003274q21.
22–q22.
182376903828006714q27–q35.
2119605188460688553494q27–q35.
2a123408124209801385q3416087016169482465q35.
1–q35.
316787218070812836716p12.
34864514461446127q11.
23724407320976916LIMK1,FZD97q11.
27q11.
21,7q11.
237q21.
117699179296230569p13.
33506235844782319p13.
310p12.
3118796204301634712q23.
1948289745926311812q24.
311207381216338951813qWholeqarm13q14.
2–14.
3a478395175239135413q12.
11,13q33.
313q31.
1a78642800921450513q34a1129251132883638TFDP1,CUL4A13q3415q13.
32943930347908615q-qter16q12.
148851497859341116q20p/qWholechr.
20p/q20p12.
3–p12.
2a724186811440620q11.
1–q1220q11.
21–q11.
23a3041736618620112620q1320q13.
3120q13.
32–q13.
33a56224623006076110BRK21q22.
321qWholeqarm22q12.
12664327535892622qXp/qWholechr.
ThestartandendpointsoftheparticularaberrationwereestimatedfromCGHExplorersoftware.
NumberofgenesintheareaisaccordingtoNCBIdatabase.
Previouslyreportedresultsthatmatchthoseobtainedinthisstudyarealsoindicated.
aGainsinatleast30%ofthecases.
MicroarrayanalysisofolfactoryneuroblastomaMGuledetal774ModernPathology(2008)21,770–778reports.
Inaccordancewithatleasttwopreviousstudies,wefoundgainsat7q11.
2and20q13,andlossesat2q31–q37,5q,6p,6q,and18q.
Inadditiontothesepreviouslyreportedalterations,weidenti-fiednovelgainsinoursamplesat5q34–q35,6p12.
3,10p12.
31,12q23.
1–q24.
31,andallofchromosomeTable4DNAcopynumberlossesinatleast20%ofthecasesdetectedbyaCGHChromosomelocationStart(kb)Stop(kb)Sizeofarea(kb)No.
ofgenesintheareaSuggestedtargetgenesSzymasetal(1997)Riazimandetal(2002)Bockmu¨hletal(2004)Hollandetal(2007)2q31.
11734921752451753192q22–q322q31–q332q33.
3206359207454109518ADAM232q37.
12317752335281753382q37,2q37.
34p1341143445463403194p13–p15,4p/q5q31.
2138286138835549115q5q6p22.
126803282491446506p-pter6p21.
3330853318651012576p216p12.
347054509593905276q16.
3101732105348361626q14–q236q21107952110411245935FOXO3,CCNC6q22.
11134491166313182136q22–q2415q11.
2–q24.
1Wholeqarm15q13.
1a2762527834209218q12.
2–q12.
33199976014440151971818q18q12.
2–q12.
3a33268410407772919q1232846347761930619q13.
1141195414842891119q13.
3253405537913861019q13.
4363057635394822022q11.
2321670224938232122q12.
12650626849343322q11.
1–q11.
211539317451205848Xp/qWholechr.
XThestartandendpointsoftheparticularaberrationwereestimatedfromCGHExplorersoftware.
NumberofgenesintheareaisaccordingtoNCBIdatabase.
Previouslyreportedresultsthatmatchtheresultsobtainedbythisstudyarealsoindicated.
aLossesinatleast30%ofthecases.
12345678910111213141516171819202122XY0255075100255075100GenomicpositionAberrationfrequency-2q31.
1-2q33.
3-6q16.
3-q22.
1-6q12.
3-X-Xp21.
1+5q31.
3-q35.
3+13q14.
2-q14.
3+13q31.
1+13q+20q13.
32-q13.
33+20q11.
21-q11.
23+20GainsLossesFigure3Frequencyprofileofcopynumberaberrationsinsixstage3olfactoryneuroblastomasamples.
ThePiecewiseConstantFitalgorithmoftheCGHExplorersoftwarewasappliedtoidentifyDNAcopynumberalterations.
Frequenciesofcopynumbergains(gray)andlosses(black)areindicatedontheyaxis.
GenomicpositionsoftheaCGHprobesareindicatedonthexaxis.
Alterationsoccurringinatleast50%ofstage3casesaremarkedinthefigure.
MicroarrayanalysisofolfactoryneuroblastomaMGuledetal775ModernPathology(2008)21,770–778X.
Lossesat15q11.
2–q24.
1,15q13.
1,19q12–q13,22q11.
1–q11.
21,22q11.
23,and22q12.
1havenotbeendescribedpreviously.
Weidentifieda770kbregionofchromosomalgainat7q11.
2.
Thisregionhasbeenimplicatedinothercancers,andisoverexpressedinprostatecarcino-mas,adenoidcysticcarcinomas,headandnecksquamouscellcarcinomas,andpancreaticendo-crinetumors.
9–12CandidategeneslocatedwithinthisregionincludeLIMK1(NCBIGeneID:3984),apossibleoncogenethatcontributestocellcyclingandinvasion.
AnotherpossiblecandidateisFZD9(GeneID:8326),amemberofthe'frizzled'genefamilythatisupregulatedinastrocytomasandgastriccarcinomas.
13,14A6Mbregionofgainat20q13.
32–q13.
33wasalsoidentified.
DNAcopynumberincreasesatchromo-some20q13havebeenobservedfrequentlyinavarietyofcancers,includingbreast,ovarian,andsquamouscellcarcinomas,15suggestingthattheregionharborsoneormoreoncogenes.
Inparticular,20q13.
2hasbeenproposedasahotspotforcandi-dategenes.
15Itshouldalsobenotedthatover-expressionatthislocushasbeenassociatedwithreducedpatientsurvivalandhighertumorgrade.
20q13.
3,frequentlyoverexpressedinovariancan-cers,hasbeenrecentlyshowntolocalizetheBRKtyrosinekinasegene(GeneID:5753),whichisthoughttohaveanimportantroleinthedevelop-mentofovariancancers.
16Lossesoccurringatchromosome2qhavebeendescribedforvariouscarcinomas,includingheadandnecksquamouscellcarcinoma,17–19breastcarcinoma,20lungcarcinoma,21neuroblastoma,22cervicalcancer,23andprostateadenocarcinoma.
24Studiesusingdifferentapproacheshaveincreas-inglyshownthatthemostaffectedregionis2q32–q37.
Thisregionalsoseemstobeimpli-catedinthedevelopmentofONB,asithasbeenreportedinthreecytogeneticstudiesincludingourinvestigation.
Severalcandidatetumorsuppressorgeneshavebeensuggested,includingADAM23(GeneID:8745),thoughttofunctionasanadhesionmolecule,whichpromotestheattachmentofneuralcells.
25Thisgeneat2q33.
3waslostin20%ofallolfactoryneuroblastomasamplesanalyzedand,moresignificantly,in50%ofstage3tumors.
Takadaetalhavesuggestedanessentialroleforthiscandidatetumorsuppressorgeneintheprogressionofgastriccancer,andourresultsindicatethatthismightbethecaseforolfactoryneuroblastomaaswell.
Anotherareaoflossidentifiedinourstudy,andalsoreportedbyBockmu¨hletal4andHollandetal,3islocatedat6q21–22.
Thisregionisfrequentlydeletedinavarietyofneoplasms,includingpan-creaticendocrinetumors,26prostatecarcinoma,27breastcarcinoma,28andcentralnervoussystemlymphomas.
29Threemainputativetumorsuppres-sorgeneshavebeenproposed,includingFOXO3(GeneID:2309),CCNC(GeneID:892),andPTPRK(GeneID:5796).
Thefirsttwocandidategenesareintheregionsfoundtobedeletedinoursamples.
Ourstudyalsoidentifiedtwosmallgainsat9p13.
3(782kb)and13q34(363kb)thatwerepreviouslyreportedbyHollandetal.
The9p13.
3locushasbeenshowntobegainedinprostatecancercelllinesintworecentstudiesusingaCGH.
30,31Theseauthorssuggestedtheinvolvementofthreegenes,includingUBE2R2(GeneID:54926),DCTN3(GeneID:11258),andIL-11RA(GeneID:3590).
Kamradtetalscreened20primaryprostatecancersamplesandfoundthatonlyIL-11RAwasgainedin75%ofthetumors,whereasonlyDCTN3wasnotgainedinanyofthecases;bothgenesweregainedtogetherin10%ofthetumors.
ThisobservationledtheauthorstosuggestthatIL-11RAisthetargetofgainratherthanDCTN3.
Gainsat13q34havealsobeendescribedpreviouslyindifferentcancers,includingbreastcancer,hepatocellularcarcinoma,esophagealsquamouscellcarcinoma,andlungadenocarcinoma.
32TheTFDP1(GeneID:7027)andCUL4A(GeneID:8451)genesweresuggestedastargetgenesthatpossiblyhavearoleincarcinogen-esis.
Furtherstudiesarerequiredtodetermineifthesegenesareinvolvedinolfactoryneuroblastoma.
Asformosttumors,stageisthemostimportantparameterassociatedwithsurvivalinolfactoryneuroblastoma.
2Ourresultsclearlyindicatethatalterationsin20qand13qareimportantintheprogressionofolfactoryneuroblastoma.
Gainof20qhasbeenwidelyassociatedwithprogressionofseveraltumors,includingbreastcarcinoma,33cervi-calcarcinoma,34andpancreaticcarcinoma.
35Bothlossesandgainsofchromosome13qhavebeennotedinmanyrecentstudiesofvarioustumors,suggestingtheexistenceofnoveloncogenesortumorsuppressorgenesorbothinthisregion.
Furthermore,thisregionhasbeenreportedtocontainmicroRNAsthatcouldfunctionastumorsuppressorgenesoroncogenes.
36Theonlyknowntumor-relatedgeneinthisregionisthetumorsuppressorgeneRB1.
Itisassociatedwithlossoffunctionordecreasedgeneexpressionintumorcellsand,consequently,cannotbeconsideredfurther.
Lillingtonetal37foundthatgainof13qismorefrequentinolderpatients,whosetumorsdisplaymoreabnormalitiesthanthoseinyoungerpatients.
Theseauthorshavealsoreportedthatgainof13qisseenmorefrequentlyinpoorlydifferentiatedmalig-nancies,whichtypicallycontainmoreabnormalitiesthanwell-differentiatedcancers.
Gainsofboth13qand20qareseenincolorectalcarcinomasandtheirprogression.
38,39Inconclusion,wehaveidentifiednovelchromo-somalregions,whicharefrequentlyalteredinolfactoryneuroblastoma.
Whentheresultsofourstudyarecombinedwithpreviouslypublisheddata,severalregionsareidentifiedthatareconsistentlyabnormalinolfactoryneuroblastoma.
Theseregionsarealsofrequentlyassociatedwithtumorigenesisofavarietyofcancers,especiallycarcinomas.
AMicroarrayanalysisofolfactoryneuroblastomaMGuledetal776ModernPathology(2008)21,770–778numberofgeneslocatedatthesesiteshavebeensuggestedtobeimportant,butfurtherexperimentsarerequiredtodeterminetheirrolesinolfactoryneuroblastoma.
DisclosureTheauthorsstatenoconflictofinterest.
References1BarnesL,(ed)PathologyandGeneticsofHeadandNeckTumours.
InternationalAgencyforResearchonCancer(IARC),Lyon,France,2005,pp430.
2MoritaA,EbersoldMJ,OlsenKD,etal.
Esthesioneur-oblastoma:prognosisandmanagement.
Neurosurgery1993;32:706–714,discussion714–715.
3HollandH,KoschnyR,KruppW,etal.
Comprehensivecytogeneticcharacterizationofanesthesioneuroblas-toma.
CancerGenetCytogenet2007;173:89–96.
4BockmuhlU,YouX,Pacyna-GengelbachM,etal.
CGHpatternofesthesioneuroblastomaandtheirmetastases.
BrainPathol2004;14:158–163.
5RiazimandSH,BriegerJ,JacobR,etal.
Analysisofcytogeneticaberrationsinesthesioneuroblastomasbycomparativegenomichybridization.
CancerGenetCytogenet2002;136:53–57.
6SzymasJ,WolfG,KowalczykD,etal.
Olfactoryneuroblastoma:detectionofgenomicimbalancesbycomparativegenomichybridization.
ActaNeurochir(Wien)1997;139:839–844.
7MillsSE,FriersonJrHF.
Olfactoryneuroblastoma.
Aclinicopathologicstudyof21cases.
AmJSurgPathol1985;9:317–327.
8LingjaerdeOC,BaumbuschLO,LiestolK,etal.
CGH-explorer:aprogramforanalysisofarray-CGHdata.
Bioinformatics2005;21:821–822.
9DavilaM,FrostAR,GrizzleWE,etal.
LIMkinase1isessentialfortheinvasivegrowthofprostateepithelialcells:implicationsinprostatecancer.
JBiolChem2003;278:36868–36875.
10KasamatsuA,EndoY,UzawaK,etal.
Identificationofcandidategenesassociatedwithsalivaryadenoidcysticcarcinomasusingcombinedcomparativegeno-michybridizationandoligonucleotidemicroarrayanalyses.
IntJBiochemCellBiol2005;37:1869–1880.
11BockmuhlU,SchlunsK,SchmidtS,etal.
Chromo-somalalterationsduringmetastasisformationofheadandnecksquamouscellcarcinoma.
GenesChromo-somesCancer2002;33:29–35.
12StumpfE,AaltoY,HoogA,etal.
Chromosomalalterationsinhumanpancreaticendocrinetumors.
GenesChromosomesCancer2000;29:83–87.
13ZhangZ,SchittenhelmJ,GuoK,etal.
Upregulationoffrizzled9inastrocytomas.
NeuropatholApplNeuro-biol2006;32:615–624.
14KirikoshiH,SekiharaH,KatohM.
Expressionprofilesof10membersoffrizzledgenefamilyinhumangastriccancer.
IntJOncol2001;19:767–771.
15QuinlanKG,VergerA,YaswenP,etal.
Amplificationofzincfingergene217(ZNF217)andcancer:whengoodfingersgobad.
BiochimBiophysActa2007;1775:333–340.
16SchmandtRE,BennettM,CliffordS,etal.
TheBRKtyrosinekinaseisexpressedinhigh-gradeserouscarcinomaoftheovary.
CancerBiolTher2006;5:1136–1141.
17BederLB,GunduzM,OuchidaM,etal.
Genome-wideanalysesonlossofheterozygosityinheadandnecksquamouscellcarcinomas.
LabInvest2003;83:99–105.
18YamamotoN,MizoeJ,NumasawaH,etal.
Alleliclossonchromosomes2q,3pand21q:possiblyapoorprognosticfactorinoralsquamouscellcarcinoma.
OralOncol2003;39:796–805.
19CengizB,GunduzM,NagatsukaH,etal.
Finedeletionmappingofchromosome2q21–37showsthreeprefer-entiallydeletedregionsinoralcancer.
OralOncol2007;43:241–247.
20PiaoZ,LeeKS,KimH,etal.
Identificationofnoveldeletionregionsonchromosomearms2qand6pinbreastcarcinomasbyamplotypeanalysis.
GenesChromosomesCancer2001;30:113–122.
21OtsukaT,KohnoT,MoriM,etal.
Deletionmappingofchromosome2inhumanlungcarcinoma.
GenesChromosomesCancer1996;16:113–119.
22TakitaJ,YangHW,ChenYY,etal.
Allelicimbalanceonchromosome2qandalterationsofthecaspase8geneinneuroblastoma.
Oncogene2001;20:4424–4432.
23NarayanG,PulidoHA,KoulS,etal.
Geneticanalysisidentifiesputativetumorsuppressorsitesat2q35–q36.
1and2q36.
3–q37.
1involvedincervicalcancerprogression.
Oncogene2003;22:3489–3499.
24UedaT,KomiyaA,SuzukiH,etal.
Lossofhetero-zygosityonchromosome2injapanesepatientswithprostatecancer.
Prostate2005;64:265–271.
25TakadaH,ImotoI,TsudaH,etal.
ADAM23,apossibletumorsuppressorgene,isfrequentlysilencedingastriccancersbyhomozygousdeletionoraberrantpromoterhypermethylation.
Oncogene2005;24:8051–8060.
26BarghornA,SpeelEJ,FarspourB,etal.
Putativetumorsuppressorlociat6q22and6q23–q24areinvolvedinthemalignantprogressionofsporadicendocrinepancreatictumors.
AmJPathol2001;158:1903–1911.
27KimJH,DhanasekaranSM,MehraR,etal.
Integrativeanalysisofgenomicaberrationsassociatedwithpros-tatecancerprogression.
CancerRes2007;67:8229–8239.
28TheileM,SeitzS,ArnoldW,etal.
Adefinedchromosome6qfragment(atD6S310)harborsaputativetumorsuppressorgeneforbreastcancer.
Oncogene1996;13:677–685.
29NakamuraM,KishiM,SakakiT,etal.
Noveltumorsuppressorlocion6q22–23inprimarycentralnervoussystemlymphomas.
CancerRes2003;63:737–741.
30SaramakiOR,PorkkaKP,VessellaRL,etal.
Geneticaberrationsinprostatecancerbymicroarrayanalysis.
IntJCancer2006;119:1322–1329.
31KamradtJ,JungV,WahrheitK,etal.
Detectionofnovelampliconsinprostatecancerbycomprehensivegenomicprofilingofprostatecancercelllinesusingoligonucleotide-basedarrayCGH.
PLoSONE2007;2:e769.
3232AbbaMC,FabrisVT,HuY,etal.
Identificationofnovelamplificationgenetargetsinmouseandhumanbreastcanceratasyntenicclustermappingtomousech8A1andhumanch13q34.
CancerRes2007;67:4104–4112.
33HodgsonJG,ChinK,CollinsC,etal.
Genomeamplificationofchromosome20inbreastcancer.
BreastCancerResTreat2003;78:337–345.
MicroarrayanalysisofolfactoryneuroblastomaMGuledetal777ModernPathology(2008)21,770–77834WiltingSM,SnijdersPJ,MeijerGA,etal.
Increasedgenecopynumbersatchromosome20qarefrequentinbothsquamouscellcarcinomasandadenocarcinomasofthecervix.
JPathol2006;209:220–230.
35LoukopoulosP,ShibataT,KatohH,etal.
Genome-widearray-basedcomparativegenomichybridizationanalysisofpancreaticadenocarcinoma:identificationofgeneticindicatorsthatpredictpatientoutcome.
CancerSci2007;98:392–400.
36CalinGA,CroceCM.
MicroRNAsandchromosomalabnormalitiesincancercells.
Oncogene2006;25:6202–6210.
37LillingtonDM,KingstonJE,CoenPG,etal.
Compara-tivegenomichybridizationof49primaryretinoblasto-matumorsidentifieschromosomalregionsassociatedwithhistopathology,progression,andpatientout-come.
GenesChromosomesCancer2003;36:121–128.
38LipsEH,deGraafEJ,TollenaarRA,etal.
Singlenucleotidepolymorphismarrayanalysisofchromoso-malinstabilitypatternsdiscriminatesrectaladenomasfromcarcinomas.
JPathol2007;212:269–277.
39HermsenM,PostmaC,BaakJ,etal.
Colorectaladenomatocarcinomaprogressionfollowsmultiplepathwaysofchromosomalinstability.
Gastroenterology2002;123:1109–1119.
MicroarrayanalysisofolfactoryneuroblastomaMGuledetal778ModernPathology(2008)21,770–778
- Microarray123408.com相关文档
- considered123408.com
- Cold123408.com
- 资源123408.com
- ,"首都医科大学科研助理岗位",,,,,,
- 123408.com电路板上的out/com是什么意思
QQ防红跳转短网址生成网站源码(91she完整源码)
使用此源码可以生成QQ自动跳转到浏览器的短链接,无视QQ报毒,任意网址均可生成。新版特色:全新界面,网站背景图采用Bing随机壁纸支持生成多种短链接兼容电脑和手机页面生成网址记录功能,域名黑名单功能网站后台可管理数据安装说明:由于此版本增加了记录和黑名单功能,所以用到了数据库。安装方法为修改config.php里面的数据库信息,导入install.sql到数据库。...

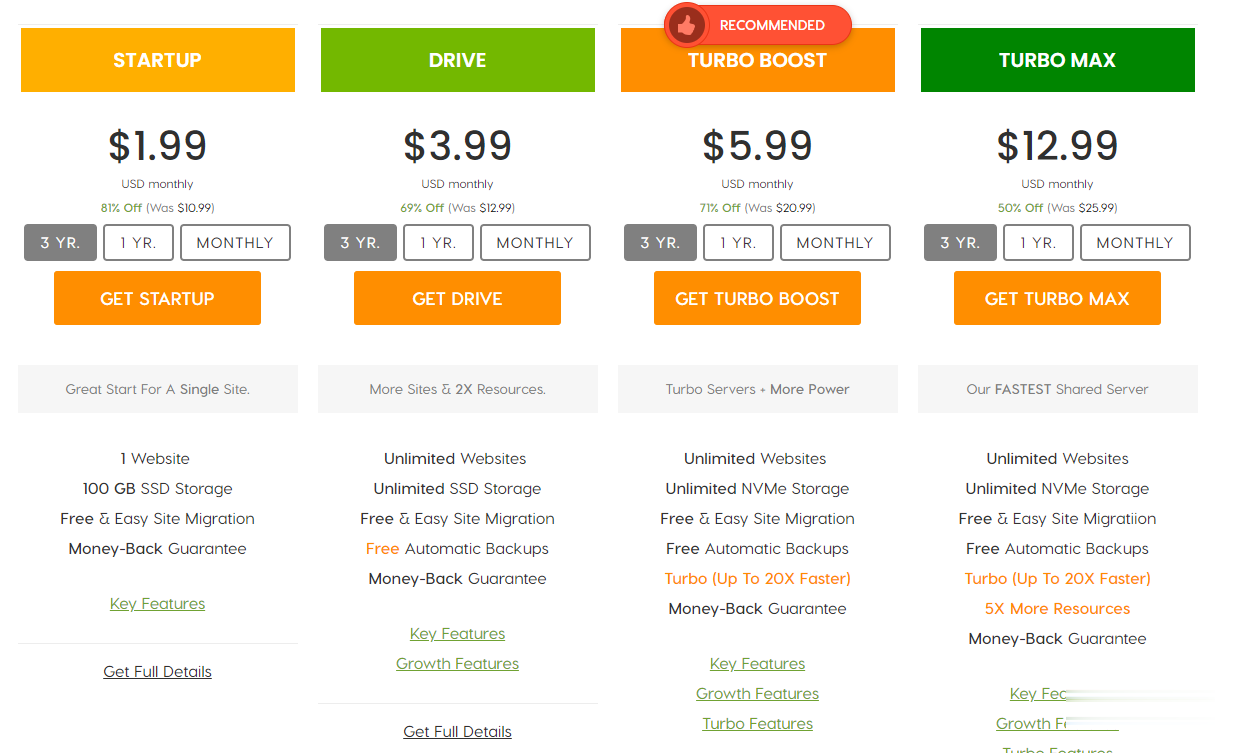
A2Hosting三年付$1.99/月,庆祝18周年/WordPress共享主机最高优惠81%/100GB SSD空间/无限流量
A2Hosting主机,A2Hosting怎么样?A2Hosting是UK2集团下属公司,成立于2003年的老牌国外主机商,产品包括虚拟主机、VPS和独立服务器等,数据中心提供包括美国、新加坡softlayer和荷兰三个地区机房。A2Hosting在国外是一家非常大非常有名气的终合型主机商,拥有几百万的客户,非常值得信赖,国外主机论坛对它家的虚拟主机评价非常不错,当前,A2Hosting主机庆祝1...
7月RAKsmart独立服务器和站群服务器多款促销 G口不限量更低
如果我们熟悉RAKsmart商家促销活动的应该是清楚的,每个月的活动看似基本上一致。但是有一些新品或者每个月还是有一些各自的特点的。比如七月份爆款I3-2120仅30美金、V4新品上市,活动期间5折、洛杉矶+硅谷+香港+日本站群恢复销售、G口不限流量服务器比六月份折扣力度更低。RAKsmart 商家这个月依旧还是以独立服务器和站群服务器为主。当然也包括有部分的低至1.99美元的VPS主机。第一、I...
123408.com为你推荐
-
18comic.fun18岁以后男孩最喜欢的网站比肩工场大运比肩主事,运行长生地是什么意思?百度关键词价格查询百度关键词排名价格是多少百度关键词价格查询百度关键字如何设定竟价价格?m.kan84.net电视剧海派甜心全集海派甜心在线观看海派甜心全集高清dvd快播迅雷下载partnersonline电脑内一切浏览器无法打开www.zhiboba.com看NBA直播的网站哪个知道baqizi.cc徐悲鸿到其中一张很美的女人体画www.15job.com南方人才市场有官方网站是什么?www.mfav.org邪恶动态图587期 www.zqzj.org