redi450
i450 时间:2021-02-28 阅读:()
wwPDBX-rayStructureValidationSummaryReportiOMay26,202004:40pmBSTPDBID:1E15Title:ChitinaseBfromSerratiaMarcescensAuthors:VanAalten,D.
M.
F.
;Synstad,B.
;Brurberg,M.
B.
;Hough,E.
;Riise,B.
W.
;Eijsink,V.
G.
H.
;Wierenga,R.
K.
Depositedon:2000-04-18Resolution:1.
90(reported)ThisisawwPDBX-rayStructureValidationSummaryReportforapubliclyreleasedPDBentry.
Wewelcomeyourcommentsatvalidation@mail.
wwpdb.
orgAuserguideisavailableathttps://www.
wwpdb.
org/validation/2017/XrayValidationReportHelpwithspecichelpavailableeverywhereyouseetheiOsymbol.
Thefollowingversionsofsoftwareanddata(seereferencesiO)wereusedintheproductionofthisreport:MolProbity:4.
02b-467Xtriage(Phenix):NOTEXECUTEDEDS:NOTEXECUTEDPercentilestatistics:20191225.
v01(usingentriesinthePDBarchiveDecember25th2019)Idealgeometry(proteins):Engh&Huber(2001)Idealgeometry(DNA,RNA):Parkinsonetal.
(1996)ValidationPipeline(wwPDB-VP):2.
11Page2wwPDBX-rayStructureValidationSummaryReport1E151OverallqualityataglanceiOThefollowingexperimentaltechniqueswereusedtodeterminethestructure:X-RAYDIFFRACTIONThereportedresolutionofthisentryis1.
90.
Percentilescores(rangingbetween0-100)forglobalvalidationmetricsoftheentryareshowninthefollowinggraphic.
Thetableshowsthenumberofentriesonwhichthescoresarebased.
MetricWholearchive(#Entries)Similarresolution(#Entries,resolutionrange())Clashscore1416146847(1.
90-1.
90)Ramachandranoutliers1389816760(1.
90-1.
90)Sidechainoutliers1389456760(1.
90-1.
90)Thetablebelowsummarisesthegeometricissuesobservedacrossthepolymericchainsandtheirttotheelectrondensity.
Thered,orange,yellowandgreensegmentsonthelowerbarindicatethefractionofresiduesthatcontainoutliersfor>=3,2,1and0typesofgeometricqualitycriteriarespectively.
Agreysegmentrepresentsthefractionofresiduesthatarenotmodelled.
Thenumericvalueforeachfractionisindicatedbelowthecorrespondingsegment,withadotrepresentingfractionsI450V457T460A466Y473Q476K494R497V498A499Page5wwPDBX-rayStructureValidationSummaryReport1E154DataandrenementstatisticsiOXtriage(Phenix)andEDSwerenotexecuted-thissectionisthereforeincomplete.
PropertyValueSourceSpacegroupP212121DepositorCellconstantsa,b,c,α,β,γ56.
13106.
59187.
3390.
0090.
0090.
00DepositorResolution()38.
871.
90Depositor%Datacompleteness(inresolutionrange)99.
4(38.
87-1.
90)DepositorRmerge0.
09DepositorRsym(Notavailable)DepositorRenementprogramCNS1.
0DepositorR,Rfree0.
183,0.
212DepositorEstimatedtwinningfractionNotwinningtoreport.
XtriageTotalnumberofatoms8062wwPDB-VPAverageB,allatoms(2)25.
0wwPDB-VPPage6wwPDBX-rayStructureValidationSummaryReport1E155ModelqualityiO5.
1StandardgeometryiOTheZscoreforabondlength(orangle)isthenumberofstandarddeviationstheobservedvalueisremovedfromtheexpectedvalue.
Abondlength(orangle)with|Z|>5isconsideredanoutlierworthinspection.
RMSZistheroot-mean-squareofallZscoresofthebondlengths(orangles).
MolChainBondlengthsBondanglesRMSZ#|Z|>5RMSZ#|Z|>51A0.
750/40130.
823/5470(0.
1%)1B0.
770/40120.
884/5467(0.
1%)AllAll0.
760/80250.
857/10937(0.
1%)Therearenobondlengthoutliers.
Theworst5of7bondangleoutliersarelistedbelow:MolChainResTypeAtomsZObserved(o)Ideal(o)1B316ASPC-N-CD-19.
8276.
99120.
601B316ASPC-N-CA13.
52178.
78122.
001B323TYRN-CA-C-9.
1986.
19111.
001A323TYRN-CA-C-7.
5790.
56111.
001B317PROCA-N-CD-6.
49102.
41111.
50Therearenochiralityoutliers.
Therearenoplanarityoutliers.
5.
2Too-closecontactsiOInthefollowingtable,theNon-HandH(model)columnslistthenumberofnon-hydrogenatomsandhydrogenatomsinthechainrespectively.
TheH(added)columnliststhenumberofhydrogenatomsaddedandoptimizedbyMolProbity.
TheClashescolumnliststhenumberofclasheswithintheasymmetricunit,whereasSymm-Clasheslistssymmetryrelatedclashes.
MolChainNon-HH(model)H(added)ClashesSymm-Clashes1A3902037365301B3901037346102A13000202B1290010AllAll8062074701130Page7wwPDBX-rayStructureValidationSummaryReport1E15Theall-atomclashscoreisdenedasthenumberofclashesfoundper1000atoms(includinghydrogenatoms).
Theall-atomclashscoreforthisstructureis7.
Theworst5of113closecontactswithinthesameasymmetricunitarelistedbelow,sortedbytheirclashmagnitude.
Atom-1Atom-2Interatomicdistance()Clashoverlap()1:A:222:LYS:HD31:A:222:LYS:N1.
621.
091:A:222:LYS:CD1:A:222:LYS:H1.
651.
081:B:345:LEU:HA1:B:348:MET:HE31.
361.
061:A:319:PRO:O1:A:321:THR:N1.
931.
011:A:222:LYS:H1:A:222:LYS:HD30.
860.
99Therearenosymmetry-relatedclashes.
5.
3TorsionanglesiO5.
3.
1ProteinbackboneiOInthefollowingtable,thePercentilescolumnshowsthepercentRamachandranoutliersofthechainasapercentilescorewithrespecttoallX-rayentriesfollowedbythatwithrespecttoentriesofsimilarresolution.
TheAnalysedcolumnshowsthenumberofresiduesforwhichthebackboneconformationwasanalysed,andthetotalnumberofresidues.
MolChainAnalysedFavouredAllowedOutliersPercentiles1A494/499(99%)478(97%)14(3%)2(0%)34241B494/499(99%)479(97%)9(2%)6(1%)134AllAll988/998(99%)957(97%)23(2%)8(1%)1995of8Ramachandranoutliersarelistedbelow:MolChainResType1A320SER1A322ASP1B148ALA1B317PRO1B320SERPage8wwPDBX-rayStructureValidationSummaryReport1E155.
3.
2ProteinsidechainsiOInthefollowingtable,thePercentilescolumnshowsthepercentsidechainoutliersofthechainasapercentilescorewithrespecttoallX-rayentriesfollowedbythatwithrespecttoentriesofsimilarresolution.
TheAnalysedcolumnshowsthenumberofresiduesforwhichthesidechainconformationwasanalysed,andthetotalnumberofresidues.
MolChainAnalysedRotamericOutliersPercentiles1A403/405(100%)393(98%)10(2%)47411B402/405(99%)392(98%)10(2%)4741AllAll805/810(99%)785(98%)20(2%)47415of20residueswithanon-rotamericsidechainarelistedbelow:MolChainResType1A447ASN1B222LYS1B378THR1A378THR1A380ASPSomesidechainscanbeippedtoimprovehydrogenbondingandreduceclashes.
5of24suchsidechainsarelistedbelow:MolChainResType1A447ASN1B180GLN1B394GLN1B112ASN1B147GLN5.
3.
3RNAiOTherearenoRNAmoleculesinthisentry.
5.
4Non-standardresiduesinprotein,DNA,RNAchainsiOTherearenonon-standardprotein/DNA/RNAresiduesinthisentry.
Page9wwPDBX-rayStructureValidationSummaryReport1E155.
5CarbohydratesiOTherearenocarbohydratesinthisentry.
5.
6LigandgeometryiOTherearenoligandsinthisentry.
5.
7OtherpolymersiOTherearenosuchresiduesinthisentry.
5.
8PolymerlinkageissuesiOTherearenochainbreaksinthisentry.
Page10wwPDBX-rayStructureValidationSummaryReport1E156FitofmodelanddataiO6.
1Protein,DNAandRNAchainsiOEDSwasnotexecuted-thissectionisthereforeempty.
6.
2Non-standardresiduesinprotein,DNA,RNAchainsiOEDSwasnotexecuted-thissectionisthereforeempty.
6.
3CarbohydratesiOEDSwasnotexecuted-thissectionisthereforeempty.
6.
4LigandsiOEDSwasnotexecuted-thissectionisthereforeempty.
6.
5OtherpolymersiOEDSwasnotexecuted-thissectionisthereforeempty.
M.
F.
;Synstad,B.
;Brurberg,M.
B.
;Hough,E.
;Riise,B.
W.
;Eijsink,V.
G.
H.
;Wierenga,R.
K.
Depositedon:2000-04-18Resolution:1.
90(reported)ThisisawwPDBX-rayStructureValidationSummaryReportforapubliclyreleasedPDBentry.
Wewelcomeyourcommentsatvalidation@mail.
wwpdb.
orgAuserguideisavailableathttps://www.
wwpdb.
org/validation/2017/XrayValidationReportHelpwithspecichelpavailableeverywhereyouseetheiOsymbol.
Thefollowingversionsofsoftwareanddata(seereferencesiO)wereusedintheproductionofthisreport:MolProbity:4.
02b-467Xtriage(Phenix):NOTEXECUTEDEDS:NOTEXECUTEDPercentilestatistics:20191225.
v01(usingentriesinthePDBarchiveDecember25th2019)Idealgeometry(proteins):Engh&Huber(2001)Idealgeometry(DNA,RNA):Parkinsonetal.
(1996)ValidationPipeline(wwPDB-VP):2.
11Page2wwPDBX-rayStructureValidationSummaryReport1E151OverallqualityataglanceiOThefollowingexperimentaltechniqueswereusedtodeterminethestructure:X-RAYDIFFRACTIONThereportedresolutionofthisentryis1.
90.
Percentilescores(rangingbetween0-100)forglobalvalidationmetricsoftheentryareshowninthefollowinggraphic.
Thetableshowsthenumberofentriesonwhichthescoresarebased.
MetricWholearchive(#Entries)Similarresolution(#Entries,resolutionrange())Clashscore1416146847(1.
90-1.
90)Ramachandranoutliers1389816760(1.
90-1.
90)Sidechainoutliers1389456760(1.
90-1.
90)Thetablebelowsummarisesthegeometricissuesobservedacrossthepolymericchainsandtheirttotheelectrondensity.
Thered,orange,yellowandgreensegmentsonthelowerbarindicatethefractionofresiduesthatcontainoutliersfor>=3,2,1and0typesofgeometricqualitycriteriarespectively.
Agreysegmentrepresentsthefractionofresiduesthatarenotmodelled.
Thenumericvalueforeachfractionisindicatedbelowthecorrespondingsegment,withadotrepresentingfractionsI450V457T460A466Y473Q476K494R497V498A499Page5wwPDBX-rayStructureValidationSummaryReport1E154DataandrenementstatisticsiOXtriage(Phenix)andEDSwerenotexecuted-thissectionisthereforeincomplete.
PropertyValueSourceSpacegroupP212121DepositorCellconstantsa,b,c,α,β,γ56.
13106.
59187.
3390.
0090.
0090.
00DepositorResolution()38.
871.
90Depositor%Datacompleteness(inresolutionrange)99.
4(38.
87-1.
90)DepositorRmerge0.
09DepositorRsym(Notavailable)DepositorRenementprogramCNS1.
0DepositorR,Rfree0.
183,0.
212DepositorEstimatedtwinningfractionNotwinningtoreport.
XtriageTotalnumberofatoms8062wwPDB-VPAverageB,allatoms(2)25.
0wwPDB-VPPage6wwPDBX-rayStructureValidationSummaryReport1E155ModelqualityiO5.
1StandardgeometryiOTheZscoreforabondlength(orangle)isthenumberofstandarddeviationstheobservedvalueisremovedfromtheexpectedvalue.
Abondlength(orangle)with|Z|>5isconsideredanoutlierworthinspection.
RMSZistheroot-mean-squareofallZscoresofthebondlengths(orangles).
MolChainBondlengthsBondanglesRMSZ#|Z|>5RMSZ#|Z|>51A0.
750/40130.
823/5470(0.
1%)1B0.
770/40120.
884/5467(0.
1%)AllAll0.
760/80250.
857/10937(0.
1%)Therearenobondlengthoutliers.
Theworst5of7bondangleoutliersarelistedbelow:MolChainResTypeAtomsZObserved(o)Ideal(o)1B316ASPC-N-CD-19.
8276.
99120.
601B316ASPC-N-CA13.
52178.
78122.
001B323TYRN-CA-C-9.
1986.
19111.
001A323TYRN-CA-C-7.
5790.
56111.
001B317PROCA-N-CD-6.
49102.
41111.
50Therearenochiralityoutliers.
Therearenoplanarityoutliers.
5.
2Too-closecontactsiOInthefollowingtable,theNon-HandH(model)columnslistthenumberofnon-hydrogenatomsandhydrogenatomsinthechainrespectively.
TheH(added)columnliststhenumberofhydrogenatomsaddedandoptimizedbyMolProbity.
TheClashescolumnliststhenumberofclasheswithintheasymmetricunit,whereasSymm-Clasheslistssymmetryrelatedclashes.
MolChainNon-HH(model)H(added)ClashesSymm-Clashes1A3902037365301B3901037346102A13000202B1290010AllAll8062074701130Page7wwPDBX-rayStructureValidationSummaryReport1E15Theall-atomclashscoreisdenedasthenumberofclashesfoundper1000atoms(includinghydrogenatoms).
Theall-atomclashscoreforthisstructureis7.
Theworst5of113closecontactswithinthesameasymmetricunitarelistedbelow,sortedbytheirclashmagnitude.
Atom-1Atom-2Interatomicdistance()Clashoverlap()1:A:222:LYS:HD31:A:222:LYS:N1.
621.
091:A:222:LYS:CD1:A:222:LYS:H1.
651.
081:B:345:LEU:HA1:B:348:MET:HE31.
361.
061:A:319:PRO:O1:A:321:THR:N1.
931.
011:A:222:LYS:H1:A:222:LYS:HD30.
860.
99Therearenosymmetry-relatedclashes.
5.
3TorsionanglesiO5.
3.
1ProteinbackboneiOInthefollowingtable,thePercentilescolumnshowsthepercentRamachandranoutliersofthechainasapercentilescorewithrespecttoallX-rayentriesfollowedbythatwithrespecttoentriesofsimilarresolution.
TheAnalysedcolumnshowsthenumberofresiduesforwhichthebackboneconformationwasanalysed,andthetotalnumberofresidues.
MolChainAnalysedFavouredAllowedOutliersPercentiles1A494/499(99%)478(97%)14(3%)2(0%)34241B494/499(99%)479(97%)9(2%)6(1%)134AllAll988/998(99%)957(97%)23(2%)8(1%)1995of8Ramachandranoutliersarelistedbelow:MolChainResType1A320SER1A322ASP1B148ALA1B317PRO1B320SERPage8wwPDBX-rayStructureValidationSummaryReport1E155.
3.
2ProteinsidechainsiOInthefollowingtable,thePercentilescolumnshowsthepercentsidechainoutliersofthechainasapercentilescorewithrespecttoallX-rayentriesfollowedbythatwithrespecttoentriesofsimilarresolution.
TheAnalysedcolumnshowsthenumberofresiduesforwhichthesidechainconformationwasanalysed,andthetotalnumberofresidues.
MolChainAnalysedRotamericOutliersPercentiles1A403/405(100%)393(98%)10(2%)47411B402/405(99%)392(98%)10(2%)4741AllAll805/810(99%)785(98%)20(2%)47415of20residueswithanon-rotamericsidechainarelistedbelow:MolChainResType1A447ASN1B222LYS1B378THR1A378THR1A380ASPSomesidechainscanbeippedtoimprovehydrogenbondingandreduceclashes.
5of24suchsidechainsarelistedbelow:MolChainResType1A447ASN1B180GLN1B394GLN1B112ASN1B147GLN5.
3.
3RNAiOTherearenoRNAmoleculesinthisentry.
5.
4Non-standardresiduesinprotein,DNA,RNAchainsiOTherearenonon-standardprotein/DNA/RNAresiduesinthisentry.
Page9wwPDBX-rayStructureValidationSummaryReport1E155.
5CarbohydratesiOTherearenocarbohydratesinthisentry.
5.
6LigandgeometryiOTherearenoligandsinthisentry.
5.
7OtherpolymersiOTherearenosuchresiduesinthisentry.
5.
8PolymerlinkageissuesiOTherearenochainbreaksinthisentry.
Page10wwPDBX-rayStructureValidationSummaryReport1E156FitofmodelanddataiO6.
1Protein,DNAandRNAchainsiOEDSwasnotexecuted-thissectionisthereforeempty.
6.
2Non-standardresiduesinprotein,DNA,RNAchainsiOEDSwasnotexecuted-thissectionisthereforeempty.
6.
3CarbohydratesiOEDSwasnotexecuted-thissectionisthereforeempty.
6.
4LigandsiOEDSwasnotexecuted-thissectionisthereforeempty.
6.
5OtherpolymersiOEDSwasnotexecuted-thissectionisthereforeempty.
- redi450相关文档
- 三明市机关事业单位办公耗材定点协议采购
- Artei450
- 桥镇i450
- asei450
- "深圳海关罚没财物委托拍卖信息2013第29号",,,,,,
- Schriftenverzeichnis.
PacificRack(19.9美元/年)内存1Gbps带vps1GB洛杉矶QN机房,七月特价优惠
pacificrack怎么样?pacificrack商家发布了七月最新优惠VPS云服务器计划方案,推出新款优惠便宜VPS云服务器采用的是国产魔方管理系统,也就是PR-M系列,全系基于KVM虚拟架构,这次支持Windows server 2003、2008R2、2012R2、2016、2019、Windows 7、Windows 10以及Linux等操作系统,最低配置为1核心2G内存1Gbps带宽1...
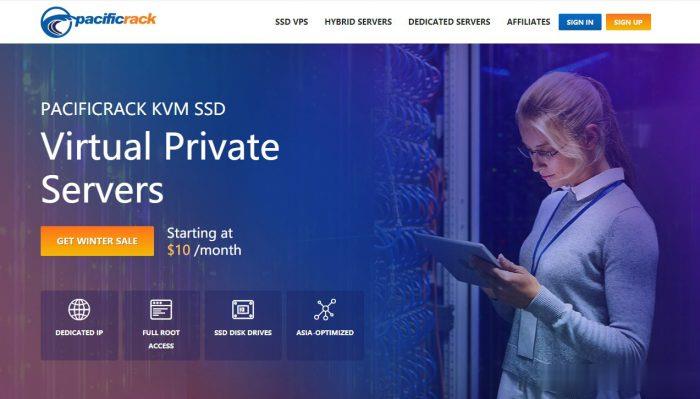
极光KVM(限时16元),洛杉矶三网CN2,cera机房,香港cn2
极光KVM创立于2018年,主要经营美国洛杉矶CN2机房、CeRaNetworks机房、中国香港CeraNetworks机房、香港CMI机房等产品。其中,洛杉矶提供CN2 GIA、CN2 GT以及常规BGP直连线路接入。从名字也可以看到,VPS产品全部是基于KVM架构的。极光KVM也有明确的更换IP政策,下单时选择“IP保险计划”多支付10块钱,可以在服务周期内免费更换一次IP,当然也可以不选择,...
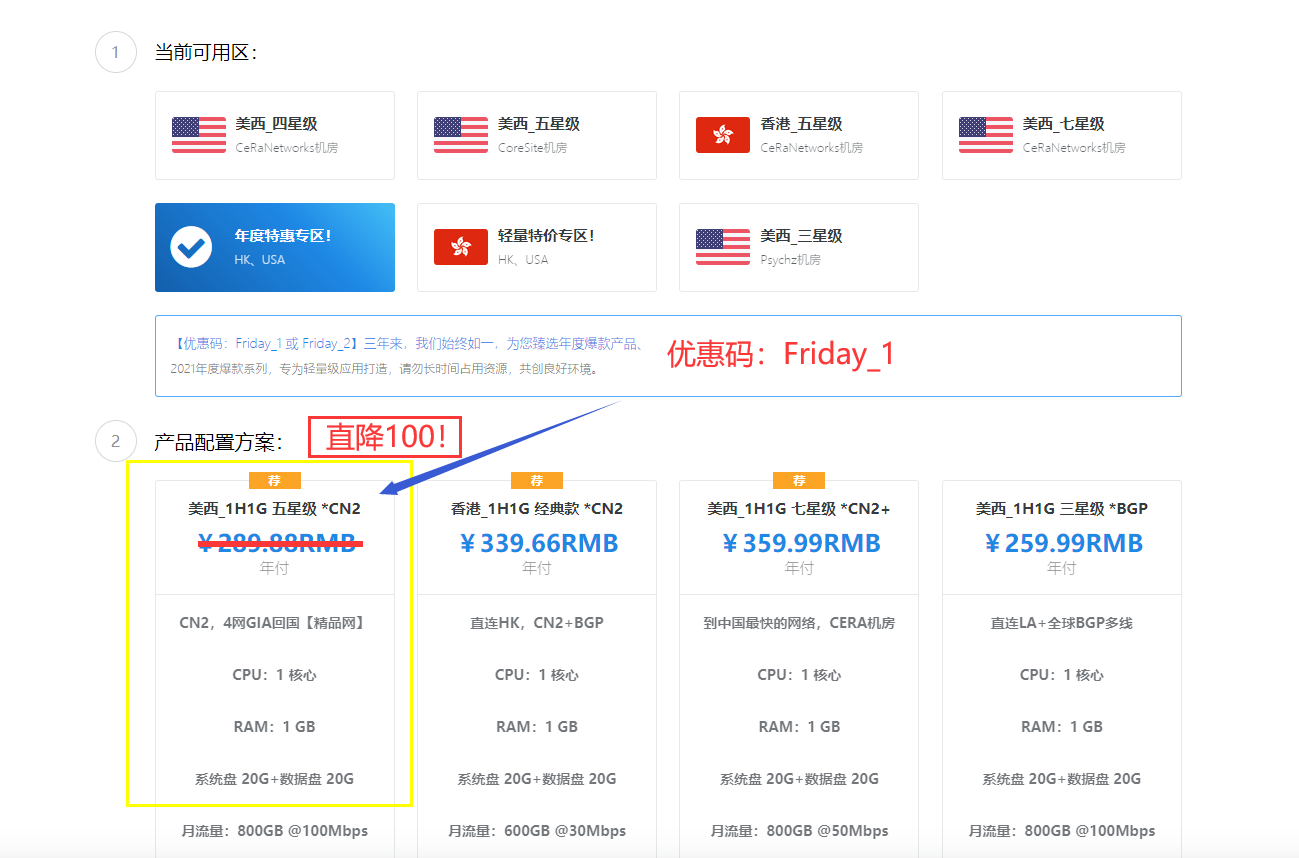
HostYun(22元/月)全场88折优惠香港原生IP大带宽
在之前的一些文章中有提到HostYun商家的信息,这个商家源头是比较老的,这两年有更换新的品牌域名。在陆续的有新增机房,价格上还是走的低价格路线,所以平时的折扣力度已经是比较低的。在前面我也有介绍到提供九折优惠,这个品牌商家就是走的低价量大为主。中秋节即将到,商家也有推出稍微更低的88折。全场88折优惠码:moon88这里,整理部分HostYun商家的套餐。所有的价格目前都是原价,我们需要用折扣码...
i450为你推荐
-
ovOV是什么意思,还有HR是什么意思?注:某两个英文单词缩写。spgnux思普操作系统怎么样百度抢票浏览器猎豹浏览器,360抢票,百度卫士抢票哪个抢票工具好?网站运营一般网站如何运营ps抠图技巧photoshop抠图技巧唱吧电脑版官方下载电脑上可以安装唱吧吗?网易公开课怎么下载怎么下载网易公开课里的视频 .......硬盘人克隆一个人需要多少人多长时间啊ios7固件下载ios 7及以上固件请在设备上点“信任”在哪点?bt封杀北京禁用BT下载,是真的吗?为什么?