arabinosewwww.youjizz.com
wwww.youjizz.com 时间:2021-04-07 阅读:()
RESEARCHOpenAccessIsolation,identificationandcharacterizationofBurkholderiapseudomalleifromsoilofcoastalregionofIndiaArchanaPrakash1,DuraipandianThavaselvam1*,AshuKumar1,AjithKumar2,SoniaArora1,SapanaTiwari1,AnitaBarua1andKannusamySathyaseelan1AbstractMelioidosisisanemerginginfectiousdiseasecausedbyafreelivingsoildwellingGram-negativebacteriumBurkholderiapseudomallei.
ThediseaseisendemictomostpartsofSoutheastAsiaandnorthernAustraliaandtheorganismhasbeenisolatedfrommoistsoilandwater.
InIndiaclinicalcasesarerecentlyreportedfromthestatesofTamilnadu,Kerala,Karnataka,Maharashtra,Orissa,Assam,WestBengal,PondicherryandTripura.
ThisstudyisaimedtoconfirmtheprevalenceofthisimportantbacterialspeciesinsoilsamplescollectedfromcoastalareasofTamilnadu.
FortyfivesoilsamplesfromfivedifferentsiteswerecollectedfromParangipettai,TamilnaduandscreenedforthepresenceofB.
pseudomallei.
Thestudyconfirmed4isolatesasB.
pseudomalleiwiththehelpofconventionalbacteriologicalmethodsandmolecularmethodsthatinclude;16SrDNAsequencing,B.
pseudomalleispecificPCR,fliCgeneRFLPandMALDI-TOFmassspectrometrybasedbacterialidentification.
ThisstudyrevealstheprevalenceanddistributionofB.
pseudomalleiinthesoilenvironmentincoastalareasofsouthernIndiaandfurthernecessitatesstudiesfromotherpartsofthecountry.
ItwillalsobehelpfultounderstandthedistributionofB.
pseudomalleiandtoaccessitsepidemiologicalimportance.
Keywords:Burkholderiapseudomallei;Ashdownagar;Melioidosis;Parangipettai;SoilisolateIntroductionMelioidosisiscausedbysoildwellingGram-negativebacteriumBurkholderiapseudomalleiandisanemerginginfectiousdiseaseinIndia.
ThediseaseismainlyendemicinSoutheastAsiaandnorthernAustraliawithhighestnumberofmelioidosiscasesreportedfromThailand.
Theglobaldistributionboundariesofmelioidosiscontinuetoexpandwellbeyondthetraditionallyrecognizedendemicregions(Currieetal.
,2008).
InIndia,clin-icalcaseshavebeenreportedfromstatesofTamilnadu,Kerala,Karnataka,Maharashtra,Orissa,Assam,WestBengal,PondicherryandTripura.
Burkholderiapseudo-malleihasbeenisolatedfromclinicalsampleslikeblood,sputum,pus,urine,synovial,peritonealandpericar-dialfluidsmostlyfromtertiarycarehospitalslocatedatVellore,TamilNaduandMangalore,Karnataka(Raghavanetal.
,1991;Kavithaetal.
,2008).
ThetrueincidenceofmelioidosisisnotknowninIndiaandrecentlylargernumbersofcaseshavebeenreportedfromthewesterncoastalareas(Vidyalakshmietal.
,2007).
Melioidosisisreferredtoas"agreatimitator"becauseofitswidespectrumofclinicalpresentations,rangingfrommildsubclinicalinfectiontofatalsepticaemiathatcanbechronic,localizedordisseminated.
Theinfectionoccursthroughinhalation,orskinabrasionsthatcomeincontactwithcontaminatedsoilorwater.
Diabetesisthemostcommonriskfactorthatisassociatedwiththediseaseandotherriskfactorsincludethalassaemia,alcoholismandrenalimpairment.
IsolationoftheorganismfromsoilisrequiredtodefinetheepidemiologyanddistributionofB.
pseudomallei,andtheassociatedrisktohumansandlive-stock(White,2003;Leelarasamee,2004).
EarlierstudieshaveshownthepresenceofB.
pseudomalleiintheenvir-onmentbasedoncultureofsoilandwaterfromdifferentgeographicregions,particularlyfromSoutheastAsiaandnorthernAustralia(Straussetal.
,1969).
Thedescription*Correspondence:dtselvam@drde.
drdo.
in1DivisionofMicrobiology,DefenceResearch&DevelopmentEstablishment,JhansiRoad,Gwalior474002,IndiaFulllistofauthorinformationisavailableattheendofthearticleaSpringerOpenJournal2014Prakashetal.
;licenseeSpringer.
ThisisanopenaccessarticledistributedunderthetermsoftheCreativeCommonsAttributionLicense(http://creativecommons.
org/licenses/by/2.
0),whichpermitsunrestricteduse,distribution,andreproductioninanymedium,providedtheoriginalworkisproperlycited.
Prakashetal.
SpringerPlus2014,3:438http://www.
springerplus.
com/content/3/1/438ofB.
thailandensis,anon-virulentbutcloselyrelatedspe-ciespresentinthesoil,hasmadetheisolationandcharacterizationofB.
pseudomalleifromsoilverychallen-ging(Brettetal.
,1998).
ThisspecieshassimilarcolonymorphologycharacteristicstoB.
pseudomalleionsolidmediaandbiochemicalandmoleculartechniquesareneededtodistinguishbetweenthem.
TheisolationofB.
pseudomalleifromdifferentsoildepthsandduringdifferentseasonsoftheyearhasbeenstudied,andquanti-tativecultureofB.
pseudomalleifromsoilsampleshasalsobeendoneinmanycountriespreviously(Smithetal.
,1995;Brooketal.
,1997).
RecentlyareviewfortheglobalpresenceanddistributionofB.
pseudomalleiclearlyindi-catesthatisolationofthisspeciesfromsoilhasnotbeenreportedfromIndia,despiteitsisolationfromhumancases(Limmathurotsakuletal.
,2013).
ThepresentstudywasundertakentoattempttheisolationofB.
pseudomal-leifromthecoastalricecultivatingareasofTamilNadu,Indiatoconfirmtheidentityofisolatesbyconventionalandmolecularmethods.
MaterialsandmethodsStudysiteandcollectionofsamplesThesoutheastcoastofParangipettai,DistrictCuddalore,Tamilnadu,India(11°49′Nand79°76′E)wasselec-tedasthesamplingsiteforthisstudy.
Parangipettaiis30.
3kmfromthemaincityofCuddaloreand183kmfromChennai.
Theannualaveragerainfallofthisareaisapprox.
945.
0mm,meanrelativehumidity57%andave-ragetemperaturerangebetween28°Cto40°Cinsum-merand18°Cto26°Cintheshortlivedwinterseason.
FivepaddyfieldswerechosenasstudysitesandthesamplingwasdonejustaftertherainyseasonfromJulytoSept.
2010atadepthof25to30cm.
Amongthefivepaddyfieldssites,sites1and2belongtoPonnanthittuvillage;sites3and4belongtoPinnathurvillagesituatedontheleftsideoftheVellarriverandsite5issitu-atedrightsideoftheVellarriver,neartoMutlurroad(Figure1).
Tensamplingpointswereselectedfromsite1tosite4andfivesamplingpointsfromsite5.
Soilsampleswerecollectedat100mdistanceintervalsfromeachotherinastraightline.
Approximately100gmofmoistsoilwerecollectedfromeachsamplingpointsandimmedi-atelyplacedintoasterile50mlconicalcentrifugetubes.
Thetubesweresealedtoavoidcontaminationandtrans-portedtothelaboratoryforfurtherprocessing.
SoilprocessingandisolationThesoilsamplescollectedwereprocessedfortheisola-tionofBurkholderiapseudomalleiasperthefollowingprotocol.
Briefly3gmofeachsoilsamplewasvigorouslymixedwith3mlofsteriledistilledwaterandleftforovernight.
100μloftheuppersurfaceliquidwasthentransferredinto5mlAshdownbrothwithasterilepipetteandincubatedat37°Cfor48hrs(Figure2).
Ashdownse-lectiveagarwasmodifiedfromtherecipeofAshdown(1979)asfollows:tryptone1.
5g,glycerol4ml,crystalviolet(25mg/ml)150μl,neutralred(25mg/ml)100μl,gentamicin8μg/mlfinalconcentrationfor100mlofmedium.
Afterincubation100μlofbrothwasplatedontoAshdownselectiveagarplatesandincubatedat42°C.
Theplatesincubatedforfourdayswerevisuallyinspecteddailyuntiltypicalcoloniesformedaspreviouslydescribed(Chantratitaetal.
,2007).
ThecolonieswerepurifiedbyfurthersubcultureonAshdownagartoconfirmthepurityandpreservedin30%glycerolstockat20°Cuntilfurtheruse.
BiochemicalandphenotypicconfirmationInitialscreeningofisolateswasperformedaccordingtothestandardprotocolsfollowedfortheidentificationofB.
pseudomallei(SentinelLaboratoryguidelines,2003).
Twostandardstrains,NCTC1688andNCTC10274,wereusedasreferencetypestrainsalongwiththeisolates.
ThecommerciallyavailableAPI20NE(Biomerieux)wasalsousedforthegenerationofbiochemicalprofilesofallsoilisolatesalongwithstandardstrain(Danceetal.
,1989;Amornchaietal.
,2007).
Resultswererecordedafterincu-bationof24hrto48hrat28°CandinterpretedreferringtothedatainterpretationtablefromtheAPI20NEman-ual.
Invitroantibioticsusceptibilityofisolatesforpoly-myxinB(100units/disc)andcolistin(25mcg/disc)werealsotestedonMueller–HintonagarbytheKirbyBauerdiskdiffusionmethod(CLSI,2007).
AssimilationofL-arabinosewasalsotestedconventionallyasbecauseitplaysakeyroleinthediscriminationofvirulentB.
pseudomalleifromthenonvirulentspeciesB.
thailan-densis(Wuthiekanumetal.
,1996).
ThesuspectedisolateswerefurtherscreenedforyellowhazeproductiononFrancisagarforthediscriminationofB.
pseudomalleifromB.
cepacia(Francisetal.
,2006).
MolecularconfirmationIdentificationbySpecificPCRAlleightbiochemicallyidentifiedB.
pseudomalleiiso-latesand59nonB.
pseudomalleiisolateswerefurtheridentifiedbyPCRproceduresbasedonamplificationof23SrDNAgeneandtheputativevirulentdeterminantTTSSgene(Table1).
PCRwasstandardizedwithfor-wardandreversePCRprimersandperformedinavol-umeof25μl,thereactionmixturecontaining200mMofeachdNTP,1.
5mMMgCl2,1*PCRbuffer,10pmolofeachprimer,1UofTaqDNApolymerase(Fermentas)and10ngDNA.
ThePCRcycleprotocolconsistofini-tialdenaturationat95°Cfor6minand30cyclesofdenaturationat95°Cfor1min,primerspecificannea-lingfor1minandextensionat72°Cfor2minwiththefinalextensionat72°Cfor10min.
PCRproductswerePrakashetal.
SpringerPlus2014,3:438Page2of10http://www.
springerplus.
com/content/3/1/438electrophoresedon1%agarosegelandvisualisedunderUVinanAlphaInnotechGelImager(AmershamPhar-maciaBiotech).
16SrDNAbasedphylogenicanalysis16SrDNAsequencingwasusedtoconfirmPCRidenti-fiedisolatesand16SrDNAsequenceofeachisolatewasBLASTanalysed(Geeetal.
,2003).
ThePCRreactionmixturefortheamplificationofthe16SrDNAgeneconsistedof200mMofeachdNTP,1.
5mMMgCl2,1*PCRbuffer,10pmolofeachprimer,1UofTaqDNApolymerase(Fermentas)and10ngDNA.
Thereactionwasmadeupto25μlwithsteriledistilledwaterandthecycleconsistedofinitialdenaturationat95°Cfor6minand30cyclesofdenaturationat95°Cfor1min,anneal-ingat50°Cfor1minandextensionat72°Cfor2minwiththefinalextensionat72°Cfor10min.
PCRprod-uctswereelectrophoresedon1.
0%agarosegelandvis-ualisedunderUVinageldocumentationsystemasabove.
Amplified16SrDNAPCRproductswerese-quencedbythedideoxychainterminationmethodusingtheBigdyeTerminatorv3.
1cyclesequencingkitandBigdyeXTerminatorPurificationkitinanABI10se-quencer(AppliedBiosystems).
ThederivedsequenceswerealignedusingDNASTARlasergene9CoreSuitandBLASTanalysiswasperformedforcomparisonwithotherbacterialspecies,sequencesavailableintheNCBIdatabase.
Adendogrambasedon16SrDNAsequenceArrowindicatesSiteofthesamplecollectionCirclesindicatespositivesitesCirclesindicatesnegativesitesFigure1SoilsamplingandfivestudysitesofParangipettai,Tamilnadu,Indiaindicatedbyarrowasshowninmap.
Prakashetal.
SpringerPlus2014,3:438Page3of10http://www.
springerplus.
com/content/3/1/438Table1PrimersusedinmolecularconfirmationstudyPrimernamePrimersequencesAnnealingtemperatureAmplificationsizePurpose/ReferenceVMP23-1-F5′CTTTTGGGTCATCCTRGA3′48°C1,051bpIdentification(Bauernfeindetal.
,1998)MP23-2-R5′TCCTACCATGCGAGACT3′BPTTS1-F5′CGTCTCTATACTGTCGAGCAATCG3′58°C548bpIdentification(Novaketal.
,2006)BPTTS1-R5′CGTGCACACCGGTCAGTATC3′F85′AGTTTGATCCTGGCTCAG3′50°C1,488bp16Ssequencing(Geeetal.
,2003)R14925′ACCTTGTTACGACTT3′fliC-F5′CTCGGATCCAACAGCAAC3′52°C1,167bpPCR-RFLP(Primerdesigned)fliC-RR-5′TATTGCAGGTACCTTCAG3′Figure2Schematicpresentationofisolationandidentificationprocedureusedinstudy.
Prakashetal.
SpringerPlus2014,3:438Page4of10http://www.
springerplus.
com/content/3/1/438wasalsoconstructedbytheneighborjoiningmethodusingMolecularEvolutionaryGeneticsAnalysisversion5.
0(MEGA5)analyticalsoftware(Tamuraetal.
,2011).
MALDI-TOFanalysisforbacterialidentificationEightbiochemicallyidentifiedisolateswerealsoMALDI-TOF/MSanalyzedfortheconfirmation(Table2).
FortheMALDI-TOF/MSanalysis,purifiedsinglecoloniesfroma24hrcultureofeachisolatewasdirectlydepos-itedonaMALDI-TOFMTP96targetplate(BrukerDaltonikGmbH),induplicateand1μlof96%formicacidtoeachspottoinactivatetheculture.
Thepreparationwasthenoverlaidwith1μlofmatrixsolutionthatcontainedasaturatedsolutionofα-cyano-4-hydroxycinnamicacidin50%acetonitrile,and2.
5%trifluoroaceticacid.
Atotalof18spots,twospotsfortheE.
colistandardand8*2fortheeightisolatesweremadeontargetplate.
Thismatrix-samplewascrystallizedbyair-dryingatroomtempe-raturefor5minutes.
MeasurementswereperformedwithaMicroflexIIImassspectrometer(BrukerDaltonik)equippedwitha337-nmnitrogenlaser.
Spectrawerere-cordedinthepositivelinearmode(delay,170ns;ionsource1voltage,20kV;ionsource2voltage,18.
5kV;lensvoltage,6kV;massrange,2–20kDa).
Eachspectrumwasobtainedafter240shotsinautomaticmodeatavariablelaserpower,andtheacquisitiontimerangedfrom30to60sperspot.
DatawereautomaticallyacquiredusingAutoXecutemethodofflexControlversion3.
3acquisitioncontrolsoftware.
The2firstrawspectraobtainedforeachisolatewereimportedintoBioTypersoftware,version3.
0(BrukerDaltonikGmbH),andwereanalyzedbystandardpatternmatchingagainstthe4110massspectraofdif-ferentbacterialspeciesasreferencefromtheBiotyperdatabase.
FlagellinC(fliC)genebasedrestrictionfragmentlengthanalysisRestrictionfragmentlengthanalysiswasperformedonthespecificPCR,16SrDNAsequencingandMALDI-TOF/MSconfirmedisolatestoobservethatwhethertheseisolatesbelongstosimilarstrains(cloneofasinglestrain)orbelongstodifferentstrains.
TheprimersfliCforward5′-CTCGGATCCAACAGCAAC-3′andfliCreverse5′-TATTGCAGGTACCTTCAG-3′wereusedfortheamplificationofthe1167bpfliCgene.
ThePCRamplificationwascarriedoutina50μlreactionTable2BiochemicalprofileofstandardstrainsandconfirmedsoilisolatesSubstrateStandardstrainsSoilisolatesBurkholderiapseudomalleiNTCC10274BurkholderiapseudomalleiNTCC1688DRDEBPS1001DRDEBPS1002DRDEBPS1003DRDEBPS1004NitrateTryptophanGlucose(acidification)ArginineUrea*Esculin*GelatinPNPGAssimilationofGlucoseArabinoseMannoseMannitolN-AcetylglucosamineMaltoseGluconateCaprateAdipateMalateCitratePhenylacetate*VariabletestsforB.
pseudomallei.
Prakashetal.
SpringerPlus2014,3:438Page5of10http://www.
springerplus.
com/content/3/1/438Table3BiochemicalprofilesofAshdowngrownsuspectedsoilisolatesconfirmedbyAPI20NESpeciesnameNO3/N2TRPGLUADHUREESCGELPNGGLUARAMNEMANNAGMALGNTCAPADIMLTCITPACOchrobactrumanthropi(n=3)Achromobacterxylosoxidans(n=5)Chromobacteriumviolaceum(n=9)Stenotrophomonasmaltophilia(n=10)Rizobiumradiobacter(n=1)Burkholderiacepacia(n=4)Delftiaacidovorans(n=5)Ralstoniapickettii(n=3)Enterococcusfaecalis(n=2)Brevundimonasvesicularis(n=1)Pseudomonasluteola(n=2)Comamonastestosterone(n=1)Pseudomonasaeruginosa(n=5)Acinetobacterlwoffii(n=1)Acinetobacterdentrificans(n=2)Pseudomonasmendocina(n=1)Chryseobacteriumindologenens(n=4)Figure3VMPPCRfortheconfirmationofisolates-Lane1:100bpplusMarker,Lane2:B.
pseudomalleiNTCC10274,Lane3:B.
pseudomalleiNTCC1688,Lane4:SoilisolateDRDEBPS1001,Lane5:DRDEBPS1002,Lane6:DRDEBPS1003,Lane7:DRDEBPS1004,Lane8:Negativecontrol.
Prakashetal.
SpringerPlus2014,3:438Page6of10http://www.
springerplus.
com/content/3/1/438Figure4TTSSPCRfortheconfirmationofisolates-Lane1:100bpplusMarker,Lane2:B.
pseudomalleiNTCC10274,Lane3:SoilisolateDRDEBPS1001,Lane4:DRDEBPS1002,Lane5:DRDEBPS1003,Lane6:DRDEBPS1004,Lane7:100bpplusMarker.
DRDEBPS1003DRDEBPS1004DRDEBPS1002DRDEBPS1001B.
pseudomalleiK96243B.
pseudomalleiPasteur52237B.
pseudomallei1710aB.
pseudomalleiNTCC1688B.
pseudomallei668B.
malleiATCC10399B.
malleiPRL-20B.
pseudomallei1106aB.
pseudomalleiPakistan9BUHB.
pseudomalleiMSHR346B.
oklahomensisC6786B.
glumaestrainP1-22-1B.
plantariistrainNIAES1723B.
thailandensisstrainCIP106301B.
ubonensisstrainGTC-P3-415B.
ambifariaAMMDB.
pyrrociniastrain2327B.
cepaciastrain717B.
vietnamiensisstrainLMG10929B.
cenocepaciastrainLMG16656B.
multivoransCGD2MB.
thailandensisBt4PMP6xxBTHxxBt4-3200.
005Figure5Dendogrambasedon16SrDNAsequencingconstructedbyneighborjoiningmethod(Barrepresents0.
005substitutionpersite).
SoilisolatesDRDEBPS1001,DRDEBPS1002,DRDEBPS1003andDRDEBPS1004showingproximitytoB.
pseudomalleistandardstrains.
Prakashetal.
SpringerPlus2014,3:438Page7of10http://www.
springerplus.
com/content/3/1/438volumewiththefollowingPCRconditions:initialde-naturationat95°Cfor6minand30cyclesofdenatur-ationat95°Cfor1min,annealingat52°Cfor1minandextensionat72°Cfor2minwiththefinalextensionat72°Cfor10min.
PCRproductswerepurifiedusingagelextractionpurificationkit(Qiagen)andconcentrationwasdeterminedspectrophotometrically.
Restrictiondi-gestionwasperformedwiththreerestrictionenzymesDdeI,MspIandPstI(Fermentas,fastdigest).
Restric-tiondigestionreactionmixturecontained7μlofpuri-fiedfliCPCRproductsataconc.
of100ng/μland1Uofrestrictionenzymes.
Restrictiondigestionreactionwascarriedoutin0.
5mltubesincubatedat37°Cfor2hrsandtheninactivatedbykeepingtubesinadrybathat65°Cfor10minutes.
Thendigestedproductswereelec-trophoresedon1.
8%agarosegelandvisualizedunderUVinanAlphaInnotechGelImager.
Results&DiscussionFromthe45soilsamples67isolatesweresuspectedandselectedaspossibleB.
pseudomalleibasedoncharacteris-ticcolonialmorphologyanddyeabsorptiononAshdownagarafterincubationof48hrat42°C.
Conventionalbio-chemicalprofilesofeachofthese67isolateswastestedandcomparedwiththeprofilesofB.
pseudomalleiNCTC1688andNCTC10274.
Thebiochemicalprofilesofeightisolates,namelyDRDEBPS1001,DRDEBPS1002,DRDEBPS1003,DRDEBPS1004,DRDEBPS1018,DRDEBPS1019,DRDEBPS1020andDRDEBPS1021matchedwiththebio-chemicalprofileofB.
pseudomalleiwithnegativearabin-osesugarassimilation.
Interestinglyalltheenvironmentalisolateswereabletoassimilatemaltosebutthestandardstrainscouldnotassimilatethissugar,asalsoobservedwithmostclinicalisolatesofB.
pseudomallei.
Thebio-chemicalprofileofallthese67isolateswasdeterminedbyAPI20NEandtheeightisolatesthatwereidentifiedasB.
pseudomalleibyconventionalbiochemicalassayswerealsopositivebyAPI20NE.
Theidentificationoftheremaining59isolatesbyAPI20NEisgiveninTable3.
Theantibioticsusceptibilityofthe8isolatesthatarebio-chemicallyconsistentwithB.
pseudomalleiwastestedagainstantibiotics,polymyxin(B100units/disc)andcolis-tin(25mcg/disc)andthesusceptibilitypatternwasfoundsimilartothatoftheB.
pseudomalleistandardstrains.
Theseeightisolateswerefurtherprocessedformole-cularconfirmationandonlyfourDRDEBPS1001,DRDEBPS1002,DRDEBPS1003andDRDEBPS1004werefoundpositivebyB.
pseudomalleispecificPCRforthetruncatedregionof23SrDNA(VMPPCR)andTTSSgene(TTSSPCR)withtheamplificationof1,051bpand548bprespectively(Figures3and4).
BoththesePCRswerealsoperformedwiththeDNAof59isolatesthatwerebio-chemicallygroupedasnonB.
pseudomalleiandnoampli-ficationwasobservedintheseisolatesconfirmingthebiochemicalresults.
Sequencingof16SrDNAoftheseeightisolatesconfirmedthat,thefourisolatesshowed100%homologywiththeNCBIdatabasereferencestrainsofB.
pseudomallei.
IsolatesDRDEBPS1018,DRDEBPS1019wereidentifiedasCupriavidusnecatorandisolatesDRDEBPS1020,DRDEBPS1021asEnterobactercloacae.
Thealignedpartialsequencesof16SrDNAweresub-mittedtoGenBankandaccessionnumbersJN001986,JN001987,JN001988andJN001989wereobtained.
Aphylogenetictreebasedon16SrDNAsequencesofallfourconfirmedisolateswasalsoconstructedalongwiththestandardstrainsofB.
pseudomalleiandothercloselyrelatedspecies(Figure5).
ThephylogeneticanalysisalsorevealedthatallfourisolatescloselymatchedwiththestandardstrainsofB.
pseudomalleiandwereplacedinthesameclade.
Thefourisolatesconfirmedby16SrDNAsequencingweresubjectedtoPCR-RFLPforthefliClocusandre-strictionprofilesforthreedifferentrestrictionenzymesweregenerated.
TherestrictionprofileafterdigestionwithMspIproduced443,285,134,123,99,64,19bpsizeproducts,PstIdigestionproduced681,266,220bpsizeproductsandDdeIenzymedigestionproduced628,307,123,109bpsizeproductsandtherestrictionprofileFigure6fliCgeneRestrictionpatternsofisolatesLane:M100bpplusMarker,Lane1,7,13:B.
pseudomalleiNTCC10274,Lane2,8,14:B.
pseudomalleiNTCC1688,Lane3,9,15:SoilisolateDRDEBPS1001,Lane4,10,16:DRDEBPS1002,Lane5,11,17:SoilisolateDRDEBPS1003,Lane6,12,18:SoilisolateDRDEBPS1004.
Prakashetal.
SpringerPlus2014,3:438Page8of10http://www.
springerplus.
com/content/3/1/438forallthesethreeenzymesmatchedwithB.
pseudo-malleistandardstrainsNCTC1688andNCTC10274,therebyconfirmingallfourisolatesasB.
pseudomalleiwithidenticalrestrictionprofiles(Figure6).
RecentlyMALDI-TOFmassspectrumanalysishasbeenconsideredaneasyanddiscriminatorytoolforidentificationofbacterialspecies(Listaetal.
,2011).
TheresultsofMALDI-TOFmassspectrumanalysisoftheeightsuspectedisolatesmatchedwiththatofthe16SrDNAsequenceanalysis.
ThefourisolatesDRDEBPS1001,DRDEBPS1002,DRDEBPS1003andDRDEBPS1004wereconfirmedasB.
pseudomalleionthebasisofscorevalues2.
601,2.
099,2.
362and2.
047.
TheotherfourisolateshadbeenbiochemicallysuspectedbutnotsupportedbyboththePCRswerealsoidentifiedbyMALDI-TOFspectrumanalysisasCupriavidusnecatorandEnterobactercloacaewithscorevalueof2.
112,2.
122and2.
341,2.
241respectively,confirmingtheresultsofthe16Sanalysis.
TheisolationofB.
pseudomalleifromsoilisverycom-plexasthepresenceoflargenumbersofcloselyrelatedsoilmicroflorainterfereswithitsrecoveryalthoughuseofAshdownbrothandagarfortheisolationofB.
pseu-domalleifromsoilsamplesprovidessomeselectivity.
Thisselectivitycomesfromcrystalviolet,neutralredandgentamicinpresentintheAshdownmedium.
ThevastmajorityofB.
pseudomalleiisolatesareresistanttoaminoglycosidesduetoexpressionofamultidrugeffluxpump,thusallowinguseofgentamicinforselection.
Thisstudyconfirmsthat,althoughthismediumisusefulforinitialscreening,manyotherspeciesalsogrowonthismediumthusinterferingtherecoveryofB.
pseudo-mallei.
Francisagarisamediumthatcanclearlydif-ferentiatebetweenB.
pseudomalleiandothercloselyrelatedspeciesincludingB.
cepacia(Francisetal.
,2006).
Wealsoanalyzedall8suspectedisolatesofB.
pseudo-malleionFrancisagartoobservetheyellowhazearoundcolonies24hr.
AlthoughFrancisAgarwasnotdevelopedforprimarysoilisolationofthisspeciesitcanbeusedtoconfirmsuspectedcoloniesisolatedfromAshdownagar.
Inourstudytherateofisolationwasfoundtobe5.
97%as4isolateswereconfirmedasB.
pseudomalleioutof67suspectedisolatesfromAshdownagarand2isolateseachwereobtainedfromsite1andsite2(Figure1).
InsimilarsoilisolationstudiesconductedinMalaysiaandThailand,highisolationrateswerefoundinwetricefieldsandotherclearedandcultivatedareas(Brettetal.
,1998;Smithetal.
,1995).
ThisstudyreportsthefirstisolationofB.
pseu-domalleifromsoilcollectedfrompaddyfieldsinthecoas-talregionofIndia.
Thestudysiteselectedinthisstudyfulfilledwithallcriteriaofsoilsamplingwhichwerecon-sideredasimportantfactorsintheisolationofB.
pseu-domallei.
ThepresenceofB.
pseudomalleidependsonvariousenvironmentalfactorsthatsupportthesurvivalofB.
pseudomalleiinsoil.
ThepresenceofB.
pseudomalleiinsoilishighduringwhenploughingandplantingofseedlingstakesplace(Palasatienetal.
,2008;Currieetal.
,2004).
Themaintenanceofviablebacteriainsoilsamplesduringcollection,transportandstoragebeforeprocessinginlaboratoryisalsoanimportantfactorfortheisolation.
ThepresenceofB.
pseudomalleineedstobeinitiallyde-terminedbyculturebasedmethodsinmicrobiologicalla-boratoriesthatrequireonlybasicequipmentandprovidesliveorganismsforfurtherconfirmationbyDNAbasedmolecularmethods.
Theisolatesrecoveredfromsoilneedtobecomparedwiththoserecoveredfromclinicalcasesasitprovidesvitalinformationonthepathogenicityandvirulenceofsoilisolates(Currieetal.
,2004).
Thepresenceofidenticalgeneticpatternsamongclinicalandenviron-mentalisolatessuggestsalinkbetweenthebacteriapre-sentincontaminatedsoilandtheemergenceofindigenousmelioidosis(Chenetal.
,2010).
Futurestudiesarealsorec-ommendedwithlargenumberofsamplescollectedfromdifferentgeographicalregionsofIndiatostudythepatternofdistributionofthisimportantbacterialspeciesandalsotocorrelatetheepidemiologicalrelevanceofsoilisolationtotheoccurrenceofmelioidosis.
ConclusionWereportthefirstisolationandmolecularconfirmationofB.
pseudomalleifromsoilofpaddyfieldsinthecoas-talregionofIndia.
Theseisolateswereinitiallyidentifiedbyconventionalbiochemicalmethodsandfurthercon-firmedbyadvancedmolecularbasedmethodsof16SrDNAsequencing,B.
pseudomalleispecificPCR,PCR-RFLPatfliClocusandMALDI-TOFbasedproteinprofiling.
ThisstudyconfirmsthepresenceofB.
pseudo-malleiinsoilinthecoastalregionofIndia.
TheisolationofthisimportantbacterialspeciesfromthispartofIndiashouldinitiatefurtherstudiesontheextentofenviron-mentalandclinicalimpactofmelioidosisinIndia.
CompetinginterestsTheauthorsdeclarethattheyhavenocompetinginterests.
Authors'contributionAPcarriedoutprocessingofsoilsamplesandmolecularconfirmation,DTcarriedoutdatainspection,analysisofresultsandwritingofmanuscript,AKprovidedcomputationalhelpforthealignmentandphylogenicanalysisofsequences,AKwasresponsibleforcollectionofsoilsamplesandtransportedtothelab,SA,STAB,KShelpedinexperimentsanddataanalysis.
Allauthorsreadandapprovedthefinalmanuscript.
AcknowledgementAuthorsarethankfultoDrM.
P.
Kaushik,Director,DRDEGwaliorandDr.
UrmilTutejaHeadofMicrobiologyDivision,DRDEGwaliorfortheirkindsupportsandprovidingfacilitiesforthiswork.
ThefinancialsupporttoArchanaPrakashbyUGC,Govt.
ofIndiaisalsoacknowledged.
Authordetails1DivisionofMicrobiology,DefenceResearch&DevelopmentEstablishment,JhansiRoad,Gwalior474002,India.
2CentreforAdvancedStudiesinMarineBiology,AnnamalaiUniversity,Parangipettai,TamilNadu.
Prakashetal.
SpringerPlus2014,3:438Page9of10http://www.
springerplus.
com/content/3/1/438Received:31July2013Accepted:7August2014Published:16August2014ReferencesAmornchaiP,WirongrongC,VanapornW,YuvadeeM,RattanaphoneP,BartJC,PaulNN,NguyenVC,SurasakdiW,NicholasPJ,PeacockSJ(2007)AccuracyofBurkholderiapseudomalleiidentificationusingtheAPI20NEsystemandalatexagglutinationtest.
JClinMicrobiol45:3774–3776AshdownLR(1979)AnimprovedscreeningtechniqueforisolationofPseudomonaspseudomalleifromclinicalspecimens.
Pathology11:293–297BauernfeindA,CarstenR,DetlefM,RenateJ,InesS(1998)MolecularprocedureforrapiddetectionofBurkholderiamalleiandBurkholderiapseudomallei.
JClinMicrobiol36:2737–2741BrettPJ,DeShazerD,WoodsDE(1998)Burkholderiathailandensissp.
nov.
,aBurkholderiapseudomallei-likespecies.
IntJSystBacteriol48:317–320BrookMD,CurrieB,DesmarchelierPM(1997)IsolationandidentificationofBurkholderiapseudomalleifromsoilusingselectiveculturetechniquesandthepolymerasechainreaction.
JAppMicrobiol82:589–596ChantratitaN,VanapornW,KhaemapornB,RachaneepornT,MongkolV,DirekL,CierakulW,SurasakdiW,SasithornP,WhiteNJ,NicholasPJ,PeacockSJ(2007)BiologicalrelevanceofcolonymorphologyandphenotypicswitchingbyB.
pseudomallei.
JBacteriol189:807–817ChenYS,LinHH,MuJJ,ChiangCS,ChenCH,BuuLM,LinYE,ChenYL(2010)DistributionofmelioidosiscasesandviableBurkholderiapseudomalleiinsoil:evidenceforemergingmelioidosisinTaiwan.
JClinMicrobiol48:1432–1434ClinicalLaboratoriesStandardInstituteguidelines(2007)PerformanceStandardsforAntimicrobialSusceptibilityTesting.
SeventeenthInformationalSupplement27(1):M100–S17CurrieBJ,JacupsSP,ChengAC,FisherDA,AnsteyNM,HuffamSE,HuffamSM,KrauseVL(2004)MelioidosisepidemiologyandriskfactorsfromaprospectivewholepopulationstudyinnorthernAustralia.
TropMedIntHealth9:1167–1174CurrieBJ,DanceDA,ChengAC(2008)TheglobaldistributionofB.
pseudomalleiandmelioidosis;anupdate.
TransRSocTropMedHyg102(suppl1):S1–S4DanceDA,WuthiekanunV,NaigowitP,WhiteNJ(1989)IdentificationofPseudomonaspseudomalleiinclinicalpractice:useofsimplescreeningtestsandAPI20NE.
JClinPath42:645–648FrancisA,AiyarS,YeanC,NaingL,RavichandranM(2006)AnimprovedselectiveanddifferentialmediumfortheisolationofBurkholderiapseudomalleifromclinicalspecimens.
DiaMicrobiolInfDis55:95–99GeeJE,SacchiCT,GlassMB,DeBK,WeyantRS,LevettPN,WhitneyAM,HoffmasterAR,PopovieT(2003)Useof16SrRNAgenesequencingforrapididentificationanddifferentiationofBurkholderiapseudomalleiandB.
mallei.
JClinMicrobiol41:4647–4654KavithaS,SatyaV,RaghuSK,AnanthakrishnaSB,GeorgeKV,ChiranjayM,IndiraB(2008)Melioidosis-acaseseriesfromsouthIndia.
TransRSocTropMedHyg102(suppl1):S18–S20LeelarasameeA(2004)Recentdevelopmentinmelioidosis.
CurrOpinInfectDis17:131–136LimmathurotsakulD,DanceDAB,WuthiekanunV,KaestliM,MayoM,WarnerJ,WagnerDM,TuanyokA,WertheimH,ChengTY,MukhopadhyayC,PuthuchearyS,DayNPJ,SteinmetzI,CurrieBJ,PeacockSJ(2013)SystematicreviewandconsensusguidelinesforenvironmentalsamplingofBurkholderiapseudomallei.
PLoSNeglTropDis7(3):e2105,doi:10.
1371/journal.
pntd.
0002105ListaF,ReubsaetFransAG,SantisRD,ParchenRR,deJongAdL,KieboomJ,LaakenAL,Voskamp-VisserAI,FilloS,JansenHJ,PlasJV,PaauwA(2011)ReliableidentificationatthespecieslevelofBrucellaisolateswithMALDI-TOF-MS.
BMCMicrobiol11:267NovakRT,GlassMB,GeeJE,GalD,MayoMJ,CurrieBJ,WilkinsPP(2006)Developmentandevaluationofareal-timePCRassaytargetingthetypeIIIsecretionsystemofBurkholderiapseudomallei.
JClinMicrobiol44(1):85–90PalasatienS,LertsirivorakulR,RoyrosP,WongratanacheewinS,SermswanRW(2008)SoilphysicochemicalpropertiesrelatedtothepresenceofB.
pseudomallei.
TransRSocTropMedHyg102(suppl1):S5–S9RaghavanKR,ShenoiRP,ZaerF,AiyerR,RamamoorthyP,MehtaMN(1991)MelioidosisinIndia.
IndianJPed28:184–188SentinelLaboratoryguidelinesforsuspectedagentsofbioterrorismBurkholderiamalleiandB.
pseudomallei(2003)Americansocietyformicrobiology.
Revised2March2006SmithMD,WuthiekanunV,WalshAL,WhiteNJ(1995)QuantitativerecoveryofBurkholderiapseudomalleifromsoilinThailand.
TransRSocTropMedHyg89:488–490StraussJM,GrovesMG,MariappanM,EllisonDW(1969)MelioidosisinMalaysia.
IIDistributionofPseudomonaspseudomalleiinsoilandsurfacewater.
TransRSocTropMedHyg18:698–702TamuraK,PetersonD,PetersonN,StecherG,NeiM,KumarS(2011)MEGA5:molecularevolutionarygeneticsanalysisusingmaximumlikelihood,evolutionarydistance,andmaximumparsimonymethods.
MolBiolEvol28(10):2731–2739VidyalakshmiK,ShrikalaB,BharathiB,SuchitraU(2007)Melioidosis:anunder-diagnosedentityinwesterncoastalIndia:aclinico-microbiologicalanalysis.
IndJMedMicrobiol25:245–248WhiteNJ(2003)Melioidosis.
Lancet361:1715–1722WuthiekanumV,SmithMD,DanceDAB,WalshAL,PittTL,WhiteNJ(1996)BiochemicalcharacterizationofclinicalandenvironmentalisolatesofBurkholderiapseudomallei.
JMedMicrobiol45:408–412doi:10.
1186/2193-1801-3-438Citethisarticleas:Prakashetal.
:Isolation,identificationandcharacterizationofBurkholderiapseudomalleifromsoilofcoastalregionofIndia.
SpringerPlus20143:438.
Submityourmanuscripttoajournalandbenetfrom:7Convenientonlinesubmission7Rigorouspeerreview7Immediatepublicationonacceptance7Openaccess:articlesfreelyavailableonline7Highvisibilitywithintheeld7RetainingthecopyrighttoyourarticleSubmityournextmanuscriptat7springeropen.
comPrakashetal.
SpringerPlus2014,3:438Page10of10http://www.
springerplus.
com/content/3/1/438
ThediseaseisendemictomostpartsofSoutheastAsiaandnorthernAustraliaandtheorganismhasbeenisolatedfrommoistsoilandwater.
InIndiaclinicalcasesarerecentlyreportedfromthestatesofTamilnadu,Kerala,Karnataka,Maharashtra,Orissa,Assam,WestBengal,PondicherryandTripura.
ThisstudyisaimedtoconfirmtheprevalenceofthisimportantbacterialspeciesinsoilsamplescollectedfromcoastalareasofTamilnadu.
FortyfivesoilsamplesfromfivedifferentsiteswerecollectedfromParangipettai,TamilnaduandscreenedforthepresenceofB.
pseudomallei.
Thestudyconfirmed4isolatesasB.
pseudomalleiwiththehelpofconventionalbacteriologicalmethodsandmolecularmethodsthatinclude;16SrDNAsequencing,B.
pseudomalleispecificPCR,fliCgeneRFLPandMALDI-TOFmassspectrometrybasedbacterialidentification.
ThisstudyrevealstheprevalenceanddistributionofB.
pseudomalleiinthesoilenvironmentincoastalareasofsouthernIndiaandfurthernecessitatesstudiesfromotherpartsofthecountry.
ItwillalsobehelpfultounderstandthedistributionofB.
pseudomalleiandtoaccessitsepidemiologicalimportance.
Keywords:Burkholderiapseudomallei;Ashdownagar;Melioidosis;Parangipettai;SoilisolateIntroductionMelioidosisiscausedbysoildwellingGram-negativebacteriumBurkholderiapseudomalleiandisanemerginginfectiousdiseaseinIndia.
ThediseaseismainlyendemicinSoutheastAsiaandnorthernAustraliawithhighestnumberofmelioidosiscasesreportedfromThailand.
Theglobaldistributionboundariesofmelioidosiscontinuetoexpandwellbeyondthetraditionallyrecognizedendemicregions(Currieetal.
,2008).
InIndia,clin-icalcaseshavebeenreportedfromstatesofTamilnadu,Kerala,Karnataka,Maharashtra,Orissa,Assam,WestBengal,PondicherryandTripura.
Burkholderiapseudo-malleihasbeenisolatedfromclinicalsampleslikeblood,sputum,pus,urine,synovial,peritonealandpericar-dialfluidsmostlyfromtertiarycarehospitalslocatedatVellore,TamilNaduandMangalore,Karnataka(Raghavanetal.
,1991;Kavithaetal.
,2008).
ThetrueincidenceofmelioidosisisnotknowninIndiaandrecentlylargernumbersofcaseshavebeenreportedfromthewesterncoastalareas(Vidyalakshmietal.
,2007).
Melioidosisisreferredtoas"agreatimitator"becauseofitswidespectrumofclinicalpresentations,rangingfrommildsubclinicalinfectiontofatalsepticaemiathatcanbechronic,localizedordisseminated.
Theinfectionoccursthroughinhalation,orskinabrasionsthatcomeincontactwithcontaminatedsoilorwater.
Diabetesisthemostcommonriskfactorthatisassociatedwiththediseaseandotherriskfactorsincludethalassaemia,alcoholismandrenalimpairment.
IsolationoftheorganismfromsoilisrequiredtodefinetheepidemiologyanddistributionofB.
pseudomallei,andtheassociatedrisktohumansandlive-stock(White,2003;Leelarasamee,2004).
EarlierstudieshaveshownthepresenceofB.
pseudomalleiintheenvir-onmentbasedoncultureofsoilandwaterfromdifferentgeographicregions,particularlyfromSoutheastAsiaandnorthernAustralia(Straussetal.
,1969).
Thedescription*Correspondence:dtselvam@drde.
drdo.
in1DivisionofMicrobiology,DefenceResearch&DevelopmentEstablishment,JhansiRoad,Gwalior474002,IndiaFulllistofauthorinformationisavailableattheendofthearticleaSpringerOpenJournal2014Prakashetal.
;licenseeSpringer.
ThisisanopenaccessarticledistributedunderthetermsoftheCreativeCommonsAttributionLicense(http://creativecommons.
org/licenses/by/2.
0),whichpermitsunrestricteduse,distribution,andreproductioninanymedium,providedtheoriginalworkisproperlycited.
Prakashetal.
SpringerPlus2014,3:438http://www.
springerplus.
com/content/3/1/438ofB.
thailandensis,anon-virulentbutcloselyrelatedspe-ciespresentinthesoil,hasmadetheisolationandcharacterizationofB.
pseudomalleifromsoilverychallen-ging(Brettetal.
,1998).
ThisspecieshassimilarcolonymorphologycharacteristicstoB.
pseudomalleionsolidmediaandbiochemicalandmoleculartechniquesareneededtodistinguishbetweenthem.
TheisolationofB.
pseudomalleifromdifferentsoildepthsandduringdifferentseasonsoftheyearhasbeenstudied,andquanti-tativecultureofB.
pseudomalleifromsoilsampleshasalsobeendoneinmanycountriespreviously(Smithetal.
,1995;Brooketal.
,1997).
RecentlyareviewfortheglobalpresenceanddistributionofB.
pseudomalleiclearlyindi-catesthatisolationofthisspeciesfromsoilhasnotbeenreportedfromIndia,despiteitsisolationfromhumancases(Limmathurotsakuletal.
,2013).
ThepresentstudywasundertakentoattempttheisolationofB.
pseudomal-leifromthecoastalricecultivatingareasofTamilNadu,Indiatoconfirmtheidentityofisolatesbyconventionalandmolecularmethods.
MaterialsandmethodsStudysiteandcollectionofsamplesThesoutheastcoastofParangipettai,DistrictCuddalore,Tamilnadu,India(11°49′Nand79°76′E)wasselec-tedasthesamplingsiteforthisstudy.
Parangipettaiis30.
3kmfromthemaincityofCuddaloreand183kmfromChennai.
Theannualaveragerainfallofthisareaisapprox.
945.
0mm,meanrelativehumidity57%andave-ragetemperaturerangebetween28°Cto40°Cinsum-merand18°Cto26°Cintheshortlivedwinterseason.
FivepaddyfieldswerechosenasstudysitesandthesamplingwasdonejustaftertherainyseasonfromJulytoSept.
2010atadepthof25to30cm.
Amongthefivepaddyfieldssites,sites1and2belongtoPonnanthittuvillage;sites3and4belongtoPinnathurvillagesituatedontheleftsideoftheVellarriverandsite5issitu-atedrightsideoftheVellarriver,neartoMutlurroad(Figure1).
Tensamplingpointswereselectedfromsite1tosite4andfivesamplingpointsfromsite5.
Soilsampleswerecollectedat100mdistanceintervalsfromeachotherinastraightline.
Approximately100gmofmoistsoilwerecollectedfromeachsamplingpointsandimmedi-atelyplacedintoasterile50mlconicalcentrifugetubes.
Thetubesweresealedtoavoidcontaminationandtrans-portedtothelaboratoryforfurtherprocessing.
SoilprocessingandisolationThesoilsamplescollectedwereprocessedfortheisola-tionofBurkholderiapseudomalleiasperthefollowingprotocol.
Briefly3gmofeachsoilsamplewasvigorouslymixedwith3mlofsteriledistilledwaterandleftforovernight.
100μloftheuppersurfaceliquidwasthentransferredinto5mlAshdownbrothwithasterilepipetteandincubatedat37°Cfor48hrs(Figure2).
Ashdownse-lectiveagarwasmodifiedfromtherecipeofAshdown(1979)asfollows:tryptone1.
5g,glycerol4ml,crystalviolet(25mg/ml)150μl,neutralred(25mg/ml)100μl,gentamicin8μg/mlfinalconcentrationfor100mlofmedium.
Afterincubation100μlofbrothwasplatedontoAshdownselectiveagarplatesandincubatedat42°C.
Theplatesincubatedforfourdayswerevisuallyinspecteddailyuntiltypicalcoloniesformedaspreviouslydescribed(Chantratitaetal.
,2007).
ThecolonieswerepurifiedbyfurthersubcultureonAshdownagartoconfirmthepurityandpreservedin30%glycerolstockat20°Cuntilfurtheruse.
BiochemicalandphenotypicconfirmationInitialscreeningofisolateswasperformedaccordingtothestandardprotocolsfollowedfortheidentificationofB.
pseudomallei(SentinelLaboratoryguidelines,2003).
Twostandardstrains,NCTC1688andNCTC10274,wereusedasreferencetypestrainsalongwiththeisolates.
ThecommerciallyavailableAPI20NE(Biomerieux)wasalsousedforthegenerationofbiochemicalprofilesofallsoilisolatesalongwithstandardstrain(Danceetal.
,1989;Amornchaietal.
,2007).
Resultswererecordedafterincu-bationof24hrto48hrat28°CandinterpretedreferringtothedatainterpretationtablefromtheAPI20NEman-ual.
Invitroantibioticsusceptibilityofisolatesforpoly-myxinB(100units/disc)andcolistin(25mcg/disc)werealsotestedonMueller–HintonagarbytheKirbyBauerdiskdiffusionmethod(CLSI,2007).
AssimilationofL-arabinosewasalsotestedconventionallyasbecauseitplaysakeyroleinthediscriminationofvirulentB.
pseudomalleifromthenonvirulentspeciesB.
thailan-densis(Wuthiekanumetal.
,1996).
ThesuspectedisolateswerefurtherscreenedforyellowhazeproductiononFrancisagarforthediscriminationofB.
pseudomalleifromB.
cepacia(Francisetal.
,2006).
MolecularconfirmationIdentificationbySpecificPCRAlleightbiochemicallyidentifiedB.
pseudomalleiiso-latesand59nonB.
pseudomalleiisolateswerefurtheridentifiedbyPCRproceduresbasedonamplificationof23SrDNAgeneandtheputativevirulentdeterminantTTSSgene(Table1).
PCRwasstandardizedwithfor-wardandreversePCRprimersandperformedinavol-umeof25μl,thereactionmixturecontaining200mMofeachdNTP,1.
5mMMgCl2,1*PCRbuffer,10pmolofeachprimer,1UofTaqDNApolymerase(Fermentas)and10ngDNA.
ThePCRcycleprotocolconsistofini-tialdenaturationat95°Cfor6minand30cyclesofdenaturationat95°Cfor1min,primerspecificannea-lingfor1minandextensionat72°Cfor2minwiththefinalextensionat72°Cfor10min.
PCRproductswerePrakashetal.
SpringerPlus2014,3:438Page2of10http://www.
springerplus.
com/content/3/1/438electrophoresedon1%agarosegelandvisualisedunderUVinanAlphaInnotechGelImager(AmershamPhar-maciaBiotech).
16SrDNAbasedphylogenicanalysis16SrDNAsequencingwasusedtoconfirmPCRidenti-fiedisolatesand16SrDNAsequenceofeachisolatewasBLASTanalysed(Geeetal.
,2003).
ThePCRreactionmixturefortheamplificationofthe16SrDNAgeneconsistedof200mMofeachdNTP,1.
5mMMgCl2,1*PCRbuffer,10pmolofeachprimer,1UofTaqDNApolymerase(Fermentas)and10ngDNA.
Thereactionwasmadeupto25μlwithsteriledistilledwaterandthecycleconsistedofinitialdenaturationat95°Cfor6minand30cyclesofdenaturationat95°Cfor1min,anneal-ingat50°Cfor1minandextensionat72°Cfor2minwiththefinalextensionat72°Cfor10min.
PCRprod-uctswereelectrophoresedon1.
0%agarosegelandvis-ualisedunderUVinageldocumentationsystemasabove.
Amplified16SrDNAPCRproductswerese-quencedbythedideoxychainterminationmethodusingtheBigdyeTerminatorv3.
1cyclesequencingkitandBigdyeXTerminatorPurificationkitinanABI10se-quencer(AppliedBiosystems).
ThederivedsequenceswerealignedusingDNASTARlasergene9CoreSuitandBLASTanalysiswasperformedforcomparisonwithotherbacterialspecies,sequencesavailableintheNCBIdatabase.
Adendogrambasedon16SrDNAsequenceArrowindicatesSiteofthesamplecollectionCirclesindicatespositivesitesCirclesindicatesnegativesitesFigure1SoilsamplingandfivestudysitesofParangipettai,Tamilnadu,Indiaindicatedbyarrowasshowninmap.
Prakashetal.
SpringerPlus2014,3:438Page3of10http://www.
springerplus.
com/content/3/1/438Table1PrimersusedinmolecularconfirmationstudyPrimernamePrimersequencesAnnealingtemperatureAmplificationsizePurpose/ReferenceVMP23-1-F5′CTTTTGGGTCATCCTRGA3′48°C1,051bpIdentification(Bauernfeindetal.
,1998)MP23-2-R5′TCCTACCATGCGAGACT3′BPTTS1-F5′CGTCTCTATACTGTCGAGCAATCG3′58°C548bpIdentification(Novaketal.
,2006)BPTTS1-R5′CGTGCACACCGGTCAGTATC3′F85′AGTTTGATCCTGGCTCAG3′50°C1,488bp16Ssequencing(Geeetal.
,2003)R14925′ACCTTGTTACGACTT3′fliC-F5′CTCGGATCCAACAGCAAC3′52°C1,167bpPCR-RFLP(Primerdesigned)fliC-RR-5′TATTGCAGGTACCTTCAG3′Figure2Schematicpresentationofisolationandidentificationprocedureusedinstudy.
Prakashetal.
SpringerPlus2014,3:438Page4of10http://www.
springerplus.
com/content/3/1/438wasalsoconstructedbytheneighborjoiningmethodusingMolecularEvolutionaryGeneticsAnalysisversion5.
0(MEGA5)analyticalsoftware(Tamuraetal.
,2011).
MALDI-TOFanalysisforbacterialidentificationEightbiochemicallyidentifiedisolateswerealsoMALDI-TOF/MSanalyzedfortheconfirmation(Table2).
FortheMALDI-TOF/MSanalysis,purifiedsinglecoloniesfroma24hrcultureofeachisolatewasdirectlydepos-itedonaMALDI-TOFMTP96targetplate(BrukerDaltonikGmbH),induplicateand1μlof96%formicacidtoeachspottoinactivatetheculture.
Thepreparationwasthenoverlaidwith1μlofmatrixsolutionthatcontainedasaturatedsolutionofα-cyano-4-hydroxycinnamicacidin50%acetonitrile,and2.
5%trifluoroaceticacid.
Atotalof18spots,twospotsfortheE.
colistandardand8*2fortheeightisolatesweremadeontargetplate.
Thismatrix-samplewascrystallizedbyair-dryingatroomtempe-raturefor5minutes.
MeasurementswereperformedwithaMicroflexIIImassspectrometer(BrukerDaltonik)equippedwitha337-nmnitrogenlaser.
Spectrawerere-cordedinthepositivelinearmode(delay,170ns;ionsource1voltage,20kV;ionsource2voltage,18.
5kV;lensvoltage,6kV;massrange,2–20kDa).
Eachspectrumwasobtainedafter240shotsinautomaticmodeatavariablelaserpower,andtheacquisitiontimerangedfrom30to60sperspot.
DatawereautomaticallyacquiredusingAutoXecutemethodofflexControlversion3.
3acquisitioncontrolsoftware.
The2firstrawspectraobtainedforeachisolatewereimportedintoBioTypersoftware,version3.
0(BrukerDaltonikGmbH),andwereanalyzedbystandardpatternmatchingagainstthe4110massspectraofdif-ferentbacterialspeciesasreferencefromtheBiotyperdatabase.
FlagellinC(fliC)genebasedrestrictionfragmentlengthanalysisRestrictionfragmentlengthanalysiswasperformedonthespecificPCR,16SrDNAsequencingandMALDI-TOF/MSconfirmedisolatestoobservethatwhethertheseisolatesbelongstosimilarstrains(cloneofasinglestrain)orbelongstodifferentstrains.
TheprimersfliCforward5′-CTCGGATCCAACAGCAAC-3′andfliCreverse5′-TATTGCAGGTACCTTCAG-3′wereusedfortheamplificationofthe1167bpfliCgene.
ThePCRamplificationwascarriedoutina50μlreactionTable2BiochemicalprofileofstandardstrainsandconfirmedsoilisolatesSubstrateStandardstrainsSoilisolatesBurkholderiapseudomalleiNTCC10274BurkholderiapseudomalleiNTCC1688DRDEBPS1001DRDEBPS1002DRDEBPS1003DRDEBPS1004NitrateTryptophanGlucose(acidification)ArginineUrea*Esculin*GelatinPNPGAssimilationofGlucoseArabinoseMannoseMannitolN-AcetylglucosamineMaltoseGluconateCaprateAdipateMalateCitratePhenylacetate*VariabletestsforB.
pseudomallei.
Prakashetal.
SpringerPlus2014,3:438Page5of10http://www.
springerplus.
com/content/3/1/438Table3BiochemicalprofilesofAshdowngrownsuspectedsoilisolatesconfirmedbyAPI20NESpeciesnameNO3/N2TRPGLUADHUREESCGELPNGGLUARAMNEMANNAGMALGNTCAPADIMLTCITPACOchrobactrumanthropi(n=3)Achromobacterxylosoxidans(n=5)Chromobacteriumviolaceum(n=9)Stenotrophomonasmaltophilia(n=10)Rizobiumradiobacter(n=1)Burkholderiacepacia(n=4)Delftiaacidovorans(n=5)Ralstoniapickettii(n=3)Enterococcusfaecalis(n=2)Brevundimonasvesicularis(n=1)Pseudomonasluteola(n=2)Comamonastestosterone(n=1)Pseudomonasaeruginosa(n=5)Acinetobacterlwoffii(n=1)Acinetobacterdentrificans(n=2)Pseudomonasmendocina(n=1)Chryseobacteriumindologenens(n=4)Figure3VMPPCRfortheconfirmationofisolates-Lane1:100bpplusMarker,Lane2:B.
pseudomalleiNTCC10274,Lane3:B.
pseudomalleiNTCC1688,Lane4:SoilisolateDRDEBPS1001,Lane5:DRDEBPS1002,Lane6:DRDEBPS1003,Lane7:DRDEBPS1004,Lane8:Negativecontrol.
Prakashetal.
SpringerPlus2014,3:438Page6of10http://www.
springerplus.
com/content/3/1/438Figure4TTSSPCRfortheconfirmationofisolates-Lane1:100bpplusMarker,Lane2:B.
pseudomalleiNTCC10274,Lane3:SoilisolateDRDEBPS1001,Lane4:DRDEBPS1002,Lane5:DRDEBPS1003,Lane6:DRDEBPS1004,Lane7:100bpplusMarker.
DRDEBPS1003DRDEBPS1004DRDEBPS1002DRDEBPS1001B.
pseudomalleiK96243B.
pseudomalleiPasteur52237B.
pseudomallei1710aB.
pseudomalleiNTCC1688B.
pseudomallei668B.
malleiATCC10399B.
malleiPRL-20B.
pseudomallei1106aB.
pseudomalleiPakistan9BUHB.
pseudomalleiMSHR346B.
oklahomensisC6786B.
glumaestrainP1-22-1B.
plantariistrainNIAES1723B.
thailandensisstrainCIP106301B.
ubonensisstrainGTC-P3-415B.
ambifariaAMMDB.
pyrrociniastrain2327B.
cepaciastrain717B.
vietnamiensisstrainLMG10929B.
cenocepaciastrainLMG16656B.
multivoransCGD2MB.
thailandensisBt4PMP6xxBTHxxBt4-3200.
005Figure5Dendogrambasedon16SrDNAsequencingconstructedbyneighborjoiningmethod(Barrepresents0.
005substitutionpersite).
SoilisolatesDRDEBPS1001,DRDEBPS1002,DRDEBPS1003andDRDEBPS1004showingproximitytoB.
pseudomalleistandardstrains.
Prakashetal.
SpringerPlus2014,3:438Page7of10http://www.
springerplus.
com/content/3/1/438volumewiththefollowingPCRconditions:initialde-naturationat95°Cfor6minand30cyclesofdenatur-ationat95°Cfor1min,annealingat52°Cfor1minandextensionat72°Cfor2minwiththefinalextensionat72°Cfor10min.
PCRproductswerepurifiedusingagelextractionpurificationkit(Qiagen)andconcentrationwasdeterminedspectrophotometrically.
Restrictiondi-gestionwasperformedwiththreerestrictionenzymesDdeI,MspIandPstI(Fermentas,fastdigest).
Restric-tiondigestionreactionmixturecontained7μlofpuri-fiedfliCPCRproductsataconc.
of100ng/μland1Uofrestrictionenzymes.
Restrictiondigestionreactionwascarriedoutin0.
5mltubesincubatedat37°Cfor2hrsandtheninactivatedbykeepingtubesinadrybathat65°Cfor10minutes.
Thendigestedproductswereelec-trophoresedon1.
8%agarosegelandvisualizedunderUVinanAlphaInnotechGelImager.
Results&DiscussionFromthe45soilsamples67isolatesweresuspectedandselectedaspossibleB.
pseudomalleibasedoncharacteris-ticcolonialmorphologyanddyeabsorptiononAshdownagarafterincubationof48hrat42°C.
Conventionalbio-chemicalprofilesofeachofthese67isolateswastestedandcomparedwiththeprofilesofB.
pseudomalleiNCTC1688andNCTC10274.
Thebiochemicalprofilesofeightisolates,namelyDRDEBPS1001,DRDEBPS1002,DRDEBPS1003,DRDEBPS1004,DRDEBPS1018,DRDEBPS1019,DRDEBPS1020andDRDEBPS1021matchedwiththebio-chemicalprofileofB.
pseudomalleiwithnegativearabin-osesugarassimilation.
Interestinglyalltheenvironmentalisolateswereabletoassimilatemaltosebutthestandardstrainscouldnotassimilatethissugar,asalsoobservedwithmostclinicalisolatesofB.
pseudomallei.
Thebio-chemicalprofileofallthese67isolateswasdeterminedbyAPI20NEandtheeightisolatesthatwereidentifiedasB.
pseudomalleibyconventionalbiochemicalassayswerealsopositivebyAPI20NE.
Theidentificationoftheremaining59isolatesbyAPI20NEisgiveninTable3.
Theantibioticsusceptibilityofthe8isolatesthatarebio-chemicallyconsistentwithB.
pseudomalleiwastestedagainstantibiotics,polymyxin(B100units/disc)andcolis-tin(25mcg/disc)andthesusceptibilitypatternwasfoundsimilartothatoftheB.
pseudomalleistandardstrains.
Theseeightisolateswerefurtherprocessedformole-cularconfirmationandonlyfourDRDEBPS1001,DRDEBPS1002,DRDEBPS1003andDRDEBPS1004werefoundpositivebyB.
pseudomalleispecificPCRforthetruncatedregionof23SrDNA(VMPPCR)andTTSSgene(TTSSPCR)withtheamplificationof1,051bpand548bprespectively(Figures3and4).
BoththesePCRswerealsoperformedwiththeDNAof59isolatesthatwerebio-chemicallygroupedasnonB.
pseudomalleiandnoampli-ficationwasobservedintheseisolatesconfirmingthebiochemicalresults.
Sequencingof16SrDNAoftheseeightisolatesconfirmedthat,thefourisolatesshowed100%homologywiththeNCBIdatabasereferencestrainsofB.
pseudomallei.
IsolatesDRDEBPS1018,DRDEBPS1019wereidentifiedasCupriavidusnecatorandisolatesDRDEBPS1020,DRDEBPS1021asEnterobactercloacae.
Thealignedpartialsequencesof16SrDNAweresub-mittedtoGenBankandaccessionnumbersJN001986,JN001987,JN001988andJN001989wereobtained.
Aphylogenetictreebasedon16SrDNAsequencesofallfourconfirmedisolateswasalsoconstructedalongwiththestandardstrainsofB.
pseudomalleiandothercloselyrelatedspecies(Figure5).
ThephylogeneticanalysisalsorevealedthatallfourisolatescloselymatchedwiththestandardstrainsofB.
pseudomalleiandwereplacedinthesameclade.
Thefourisolatesconfirmedby16SrDNAsequencingweresubjectedtoPCR-RFLPforthefliClocusandre-strictionprofilesforthreedifferentrestrictionenzymesweregenerated.
TherestrictionprofileafterdigestionwithMspIproduced443,285,134,123,99,64,19bpsizeproducts,PstIdigestionproduced681,266,220bpsizeproductsandDdeIenzymedigestionproduced628,307,123,109bpsizeproductsandtherestrictionprofileFigure6fliCgeneRestrictionpatternsofisolatesLane:M100bpplusMarker,Lane1,7,13:B.
pseudomalleiNTCC10274,Lane2,8,14:B.
pseudomalleiNTCC1688,Lane3,9,15:SoilisolateDRDEBPS1001,Lane4,10,16:DRDEBPS1002,Lane5,11,17:SoilisolateDRDEBPS1003,Lane6,12,18:SoilisolateDRDEBPS1004.
Prakashetal.
SpringerPlus2014,3:438Page8of10http://www.
springerplus.
com/content/3/1/438forallthesethreeenzymesmatchedwithB.
pseudo-malleistandardstrainsNCTC1688andNCTC10274,therebyconfirmingallfourisolatesasB.
pseudomalleiwithidenticalrestrictionprofiles(Figure6).
RecentlyMALDI-TOFmassspectrumanalysishasbeenconsideredaneasyanddiscriminatorytoolforidentificationofbacterialspecies(Listaetal.
,2011).
TheresultsofMALDI-TOFmassspectrumanalysisoftheeightsuspectedisolatesmatchedwiththatofthe16SrDNAsequenceanalysis.
ThefourisolatesDRDEBPS1001,DRDEBPS1002,DRDEBPS1003andDRDEBPS1004wereconfirmedasB.
pseudomalleionthebasisofscorevalues2.
601,2.
099,2.
362and2.
047.
TheotherfourisolateshadbeenbiochemicallysuspectedbutnotsupportedbyboththePCRswerealsoidentifiedbyMALDI-TOFspectrumanalysisasCupriavidusnecatorandEnterobactercloacaewithscorevalueof2.
112,2.
122and2.
341,2.
241respectively,confirmingtheresultsofthe16Sanalysis.
TheisolationofB.
pseudomalleifromsoilisverycom-plexasthepresenceoflargenumbersofcloselyrelatedsoilmicroflorainterfereswithitsrecoveryalthoughuseofAshdownbrothandagarfortheisolationofB.
pseu-domalleifromsoilsamplesprovidessomeselectivity.
Thisselectivitycomesfromcrystalviolet,neutralredandgentamicinpresentintheAshdownmedium.
ThevastmajorityofB.
pseudomalleiisolatesareresistanttoaminoglycosidesduetoexpressionofamultidrugeffluxpump,thusallowinguseofgentamicinforselection.
Thisstudyconfirmsthat,althoughthismediumisusefulforinitialscreening,manyotherspeciesalsogrowonthismediumthusinterferingtherecoveryofB.
pseudo-mallei.
Francisagarisamediumthatcanclearlydif-ferentiatebetweenB.
pseudomalleiandothercloselyrelatedspeciesincludingB.
cepacia(Francisetal.
,2006).
Wealsoanalyzedall8suspectedisolatesofB.
pseudo-malleionFrancisagartoobservetheyellowhazearoundcolonies24hr.
AlthoughFrancisAgarwasnotdevelopedforprimarysoilisolationofthisspeciesitcanbeusedtoconfirmsuspectedcoloniesisolatedfromAshdownagar.
Inourstudytherateofisolationwasfoundtobe5.
97%as4isolateswereconfirmedasB.
pseudomalleioutof67suspectedisolatesfromAshdownagarand2isolateseachwereobtainedfromsite1andsite2(Figure1).
InsimilarsoilisolationstudiesconductedinMalaysiaandThailand,highisolationrateswerefoundinwetricefieldsandotherclearedandcultivatedareas(Brettetal.
,1998;Smithetal.
,1995).
ThisstudyreportsthefirstisolationofB.
pseu-domalleifromsoilcollectedfrompaddyfieldsinthecoas-talregionofIndia.
Thestudysiteselectedinthisstudyfulfilledwithallcriteriaofsoilsamplingwhichwerecon-sideredasimportantfactorsintheisolationofB.
pseu-domallei.
ThepresenceofB.
pseudomalleidependsonvariousenvironmentalfactorsthatsupportthesurvivalofB.
pseudomalleiinsoil.
ThepresenceofB.
pseudomalleiinsoilishighduringwhenploughingandplantingofseedlingstakesplace(Palasatienetal.
,2008;Currieetal.
,2004).
Themaintenanceofviablebacteriainsoilsamplesduringcollection,transportandstoragebeforeprocessinginlaboratoryisalsoanimportantfactorfortheisolation.
ThepresenceofB.
pseudomalleineedstobeinitiallyde-terminedbyculturebasedmethodsinmicrobiologicalla-boratoriesthatrequireonlybasicequipmentandprovidesliveorganismsforfurtherconfirmationbyDNAbasedmolecularmethods.
Theisolatesrecoveredfromsoilneedtobecomparedwiththoserecoveredfromclinicalcasesasitprovidesvitalinformationonthepathogenicityandvirulenceofsoilisolates(Currieetal.
,2004).
Thepresenceofidenticalgeneticpatternsamongclinicalandenviron-mentalisolatessuggestsalinkbetweenthebacteriapre-sentincontaminatedsoilandtheemergenceofindigenousmelioidosis(Chenetal.
,2010).
Futurestudiesarealsorec-ommendedwithlargenumberofsamplescollectedfromdifferentgeographicalregionsofIndiatostudythepatternofdistributionofthisimportantbacterialspeciesandalsotocorrelatetheepidemiologicalrelevanceofsoilisolationtotheoccurrenceofmelioidosis.
ConclusionWereportthefirstisolationandmolecularconfirmationofB.
pseudomalleifromsoilofpaddyfieldsinthecoas-talregionofIndia.
Theseisolateswereinitiallyidentifiedbyconventionalbiochemicalmethodsandfurthercon-firmedbyadvancedmolecularbasedmethodsof16SrDNAsequencing,B.
pseudomalleispecificPCR,PCR-RFLPatfliClocusandMALDI-TOFbasedproteinprofiling.
ThisstudyconfirmsthepresenceofB.
pseudo-malleiinsoilinthecoastalregionofIndia.
TheisolationofthisimportantbacterialspeciesfromthispartofIndiashouldinitiatefurtherstudiesontheextentofenviron-mentalandclinicalimpactofmelioidosisinIndia.
CompetinginterestsTheauthorsdeclarethattheyhavenocompetinginterests.
Authors'contributionAPcarriedoutprocessingofsoilsamplesandmolecularconfirmation,DTcarriedoutdatainspection,analysisofresultsandwritingofmanuscript,AKprovidedcomputationalhelpforthealignmentandphylogenicanalysisofsequences,AKwasresponsibleforcollectionofsoilsamplesandtransportedtothelab,SA,STAB,KShelpedinexperimentsanddataanalysis.
Allauthorsreadandapprovedthefinalmanuscript.
AcknowledgementAuthorsarethankfultoDrM.
P.
Kaushik,Director,DRDEGwaliorandDr.
UrmilTutejaHeadofMicrobiologyDivision,DRDEGwaliorfortheirkindsupportsandprovidingfacilitiesforthiswork.
ThefinancialsupporttoArchanaPrakashbyUGC,Govt.
ofIndiaisalsoacknowledged.
Authordetails1DivisionofMicrobiology,DefenceResearch&DevelopmentEstablishment,JhansiRoad,Gwalior474002,India.
2CentreforAdvancedStudiesinMarineBiology,AnnamalaiUniversity,Parangipettai,TamilNadu.
Prakashetal.
SpringerPlus2014,3:438Page9of10http://www.
springerplus.
com/content/3/1/438Received:31July2013Accepted:7August2014Published:16August2014ReferencesAmornchaiP,WirongrongC,VanapornW,YuvadeeM,RattanaphoneP,BartJC,PaulNN,NguyenVC,SurasakdiW,NicholasPJ,PeacockSJ(2007)AccuracyofBurkholderiapseudomalleiidentificationusingtheAPI20NEsystemandalatexagglutinationtest.
JClinMicrobiol45:3774–3776AshdownLR(1979)AnimprovedscreeningtechniqueforisolationofPseudomonaspseudomalleifromclinicalspecimens.
Pathology11:293–297BauernfeindA,CarstenR,DetlefM,RenateJ,InesS(1998)MolecularprocedureforrapiddetectionofBurkholderiamalleiandBurkholderiapseudomallei.
JClinMicrobiol36:2737–2741BrettPJ,DeShazerD,WoodsDE(1998)Burkholderiathailandensissp.
nov.
,aBurkholderiapseudomallei-likespecies.
IntJSystBacteriol48:317–320BrookMD,CurrieB,DesmarchelierPM(1997)IsolationandidentificationofBurkholderiapseudomalleifromsoilusingselectiveculturetechniquesandthepolymerasechainreaction.
JAppMicrobiol82:589–596ChantratitaN,VanapornW,KhaemapornB,RachaneepornT,MongkolV,DirekL,CierakulW,SurasakdiW,SasithornP,WhiteNJ,NicholasPJ,PeacockSJ(2007)BiologicalrelevanceofcolonymorphologyandphenotypicswitchingbyB.
pseudomallei.
JBacteriol189:807–817ChenYS,LinHH,MuJJ,ChiangCS,ChenCH,BuuLM,LinYE,ChenYL(2010)DistributionofmelioidosiscasesandviableBurkholderiapseudomalleiinsoil:evidenceforemergingmelioidosisinTaiwan.
JClinMicrobiol48:1432–1434ClinicalLaboratoriesStandardInstituteguidelines(2007)PerformanceStandardsforAntimicrobialSusceptibilityTesting.
SeventeenthInformationalSupplement27(1):M100–S17CurrieBJ,JacupsSP,ChengAC,FisherDA,AnsteyNM,HuffamSE,HuffamSM,KrauseVL(2004)MelioidosisepidemiologyandriskfactorsfromaprospectivewholepopulationstudyinnorthernAustralia.
TropMedIntHealth9:1167–1174CurrieBJ,DanceDA,ChengAC(2008)TheglobaldistributionofB.
pseudomalleiandmelioidosis;anupdate.
TransRSocTropMedHyg102(suppl1):S1–S4DanceDA,WuthiekanunV,NaigowitP,WhiteNJ(1989)IdentificationofPseudomonaspseudomalleiinclinicalpractice:useofsimplescreeningtestsandAPI20NE.
JClinPath42:645–648FrancisA,AiyarS,YeanC,NaingL,RavichandranM(2006)AnimprovedselectiveanddifferentialmediumfortheisolationofBurkholderiapseudomalleifromclinicalspecimens.
DiaMicrobiolInfDis55:95–99GeeJE,SacchiCT,GlassMB,DeBK,WeyantRS,LevettPN,WhitneyAM,HoffmasterAR,PopovieT(2003)Useof16SrRNAgenesequencingforrapididentificationanddifferentiationofBurkholderiapseudomalleiandB.
mallei.
JClinMicrobiol41:4647–4654KavithaS,SatyaV,RaghuSK,AnanthakrishnaSB,GeorgeKV,ChiranjayM,IndiraB(2008)Melioidosis-acaseseriesfromsouthIndia.
TransRSocTropMedHyg102(suppl1):S18–S20LeelarasameeA(2004)Recentdevelopmentinmelioidosis.
CurrOpinInfectDis17:131–136LimmathurotsakulD,DanceDAB,WuthiekanunV,KaestliM,MayoM,WarnerJ,WagnerDM,TuanyokA,WertheimH,ChengTY,MukhopadhyayC,PuthuchearyS,DayNPJ,SteinmetzI,CurrieBJ,PeacockSJ(2013)SystematicreviewandconsensusguidelinesforenvironmentalsamplingofBurkholderiapseudomallei.
PLoSNeglTropDis7(3):e2105,doi:10.
1371/journal.
pntd.
0002105ListaF,ReubsaetFransAG,SantisRD,ParchenRR,deJongAdL,KieboomJ,LaakenAL,Voskamp-VisserAI,FilloS,JansenHJ,PlasJV,PaauwA(2011)ReliableidentificationatthespecieslevelofBrucellaisolateswithMALDI-TOF-MS.
BMCMicrobiol11:267NovakRT,GlassMB,GeeJE,GalD,MayoMJ,CurrieBJ,WilkinsPP(2006)Developmentandevaluationofareal-timePCRassaytargetingthetypeIIIsecretionsystemofBurkholderiapseudomallei.
JClinMicrobiol44(1):85–90PalasatienS,LertsirivorakulR,RoyrosP,WongratanacheewinS,SermswanRW(2008)SoilphysicochemicalpropertiesrelatedtothepresenceofB.
pseudomallei.
TransRSocTropMedHyg102(suppl1):S5–S9RaghavanKR,ShenoiRP,ZaerF,AiyerR,RamamoorthyP,MehtaMN(1991)MelioidosisinIndia.
IndianJPed28:184–188SentinelLaboratoryguidelinesforsuspectedagentsofbioterrorismBurkholderiamalleiandB.
pseudomallei(2003)Americansocietyformicrobiology.
Revised2March2006SmithMD,WuthiekanunV,WalshAL,WhiteNJ(1995)QuantitativerecoveryofBurkholderiapseudomalleifromsoilinThailand.
TransRSocTropMedHyg89:488–490StraussJM,GrovesMG,MariappanM,EllisonDW(1969)MelioidosisinMalaysia.
IIDistributionofPseudomonaspseudomalleiinsoilandsurfacewater.
TransRSocTropMedHyg18:698–702TamuraK,PetersonD,PetersonN,StecherG,NeiM,KumarS(2011)MEGA5:molecularevolutionarygeneticsanalysisusingmaximumlikelihood,evolutionarydistance,andmaximumparsimonymethods.
MolBiolEvol28(10):2731–2739VidyalakshmiK,ShrikalaB,BharathiB,SuchitraU(2007)Melioidosis:anunder-diagnosedentityinwesterncoastalIndia:aclinico-microbiologicalanalysis.
IndJMedMicrobiol25:245–248WhiteNJ(2003)Melioidosis.
Lancet361:1715–1722WuthiekanumV,SmithMD,DanceDAB,WalshAL,PittTL,WhiteNJ(1996)BiochemicalcharacterizationofclinicalandenvironmentalisolatesofBurkholderiapseudomallei.
JMedMicrobiol45:408–412doi:10.
1186/2193-1801-3-438Citethisarticleas:Prakashetal.
:Isolation,identificationandcharacterizationofBurkholderiapseudomalleifromsoilofcoastalregionofIndia.
SpringerPlus20143:438.
Submityourmanuscripttoajournalandbenetfrom:7Convenientonlinesubmission7Rigorouspeerreview7Immediatepublicationonacceptance7Openaccess:articlesfreelyavailableonline7Highvisibilitywithintheeld7RetainingthecopyrighttoyourarticleSubmityournextmanuscriptat7springeropen.
comPrakashetal.
SpringerPlus2014,3:438Page10of10http://www.
springerplus.
com/content/3/1/438
- arabinosewwww.youjizz.com相关文档
- sumouwwww.youjizz.com
- ,"MasterAuctionInventoryCatalog",,,,,,
- clientwwww.youjizz.com
- feewwww.youjizz.com
- Frostwwww.youjizz.com
- Whitewwww.youjizz.com
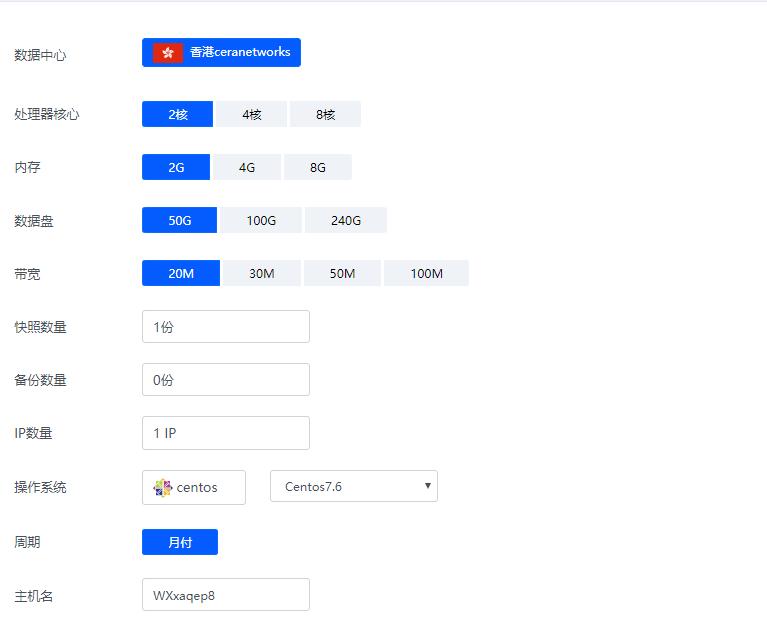
香港ceranetworks(69元/月) 2核2G 50G硬盘 20M 50M 100M 不限流量
香港ceranetworks提速啦是成立于2012年的十分老牌的一个商家这次给大家评测的是 香港ceranetworks 8核16G 100M 这款产品 提速啦老板真的是豪气每次都给高配我测试 不像别的商家每次就给1核1G,废话不多说开始跑脚本。香港ceranetworks 2核2G 50G硬盘20M 69元/月30M 99元/月50M 219元/月100M 519元/月香港ceranetwork...
piayun(pia云)240元/季起云服务器,香港限时季付活动,cn2线路,4核4G15M
pia云怎么样?pia云是一家2018的开办的国人商家,原名叫哔哔云,目前整合到了魔方云平台上,商家主要销售VPS服务,采用KVM虚拟架构 ,机房有美国洛杉矶、中国香港和深圳地区,洛杉矶为crea机房,三网回程CN2 GIA,带20G防御。目前,Pia云优惠促销,年付全场8折起,香港超极速CN2季付活动,4核4G15M云服务器仅240元/季起,香港CN2、美国三网CN2深圳BGP优质云服务器超高性...

青云互联:美国洛杉矶CN2弹性云限时八折,15元/月起,可选Windows/可自定义配置
青云互联怎么样?青云互联是一家成立于2020年6月的主机服务商,致力于为用户提供高性价比稳定快速的主机托管服务,目前提供有美国免费主机、香港主机、香港服务器、美国云服务器,让您的网站高速、稳定运行。美国cn2弹性云主机限时8折起,可选1-20个IP,仅15元/月起,附8折优惠码使用!点击进入:青云互联官方网站地址青云互联优惠码:八折优惠码:ltY8sHMh (续费同价)青云互联活动方案:美国洛杉矶...

wwww.youjizz.com为你推荐
-
杨紫别祝我生日快乐周杰伦的祝我生日快乐这首歌有什么寓意或者是在什么背景下写的同ip网站查询怎么查自己的服务器挂着哪些网站psbc.com95580是什么诈骗信息不点网址就安全吧!psbc.com怎样登录wap.psbc.com百度关键词分析如何正确分析关键词?网站检测请问,对网站进行监控检测的工具有哪些?www.119mm.comwww.993mm+com精品集!www.765.com哪里有免费的电影网站789se.comhttp://gv789.com/index.php这个网站可信吗?是真的还是假的!www.hhh258comwww.tx88d.com 有这个网站吗?