gemeizhai
meizhai 时间:2021-05-24 阅读:()
1600|OralDiseases.
2019;25:1600–1607.
wileyonlinelibrary.
com/journal/odi1|INTRODUCTIONOdontogenickeratocysts(OKCs,alsoknownaskeratocysticodon‐togenictumors)arelocallyaggressivecysticlesionsofthejawthatarethroughtoarisefromtheodontogenicepithelium(Partridge&Towers,1987).
GorlinsyndromeisanautosomaldominantdisorderpredisposingaffectedindividualstothedevelopmentofmultipleOKCs,andstudiesonpatientswithGorlinsyndromehaveinitiallyshownapivotalroleforthesonichedgehog(SHH)pathwayinOKCdevelopment.
FurtherresearchhasconfirmedthecentralroleoftheSHH/patched1(PTCH1)/smoothened(SMO)pathwayinsporadicOKCs.
PTCH1normallyactstoblocktheactivityofSMO(Stoneetal.
,1996).
Inparticular,thebindingofSHHtotheextracellularReceived:5April2019|Revised:15May2019|Accepted:28May2019DOI:10.
1111/odi.
13135ORIGINALARTICLEPTCH1alterationsarefrequentbutothergeneticalterationsarerareinsporadicodontogenickeratocystsJiafeiQu1,2|JianyunZhang1|HeyuZhang3|XuefenLi3|YingyingHong1|JiemeiZhai4|YanjinWang1|FengChen3|TiejunLi12019JohnWiley&SonsA/S.
PublishedbyJohnWiley&SonsLtd.
AllrightsreservedJiafeiQuandJianyunZhangauthorscontributedequallyandsharefirstauthorship.
1DepartmentofOralPathology,PekingUniversitySchoolandHospitalofStomatology,Beijing,China2InternationalVIPDentalClinic,TianjinStomatologicalHospital,StomatologicalHospitalofNankaiUniversity,Tianjin,China3CentralLaboratory,PekingUniversitySchoolandHospitalofStomatology,Beijing,China4DepartmentofOralPathology,SchoolofStomatology,KunmingMedicalUniversity,Kunming,ChinaCorrespondenceTiejunLi,DepartmentofOralPathology,PekingUniversitySchoolandHospitalofStomatology,22SouthZhongguancunAvenue,HaidianDistrict,Beijing100081,China.
Email:litiejun22@vip.
sina.
comFengChen,CentralLaboratory,PekingUniversitySchoolandHospitalofStomatology,22SouthZhongguancunAvenue,HaidianDistrict,Beijing100081,China.
Email:chenfeng2011@hsc.
pku.
edu.
cnFundinginformationtheBeijingNatureScienceFoundation,Grant/AwardNumber:7172238;theNationalNatureScienceFoundationofChina,Grant/AwardNumber:81030018and81671006;theFundamentalResearchFundsfortheCentralUniversities,NankaiUniversity,Grant/AwardNumber:63191118AbstractObjective:Odontogenickeratocysts(OKCs)arebenignjawlesionswithhighgrowthpotentialandpropensityforrecurrence.
OurpreviousstudyrevealedthatPTCH1mu‐tations,whichwerefrequentlydetectedinsporadicOKCs,mightbeunderestimatedduetothemaskingeffectofthestromalcomponentswithinthetestedtissues.
WeaimedtoconfirmtheseresultsinlargerscaleandfurtherpresenttheunbiasedviewofthegenomicbasisofsporadicOKCsexceptPTCH1.
Materialsandmethods:WeanalyzedPTCH1mutationsinadditional19samples.
Usingwhole‐exomesequencing(WES),wefurthercharacterizedthemutationallandscapeoffivesporadicOKCsampleslackingPTCH1mutationandlossofhet‐erozygosity(LOH).
Results:Combinedwithourpreviouslyreported19cases,thirtyof38(79%)casesharboredPTCH1mutations.
Throughwhole‐exomesequencingandintegrativeanal‐ysis,22novelmutationswereconfirmedamongfivePTCH1‐negativesamples.
NorecurrentmutationswereidentifiedintheWESsamplesandvalidationcohortof10OKCs.
Conclusions:OurdatafurtherconfirmedthefrequentPTCH1mutationandotherraregeneticalterationsinsporadicOKCs,highlightingthecentralroleofSHHsignal‐ingpathway.
InPTCH1‐negativecases,otherraremutationsscatteredinasubsetofOKCswereindependentoftheSHHpathway.
TheseresultssuggestedthatanSHHinhibitormaybeeffectivetotreatthemajorityofOKCs.
KEYWORDSPTCH1mutation,odontogenickeratocysts,whole‐exomesequencing|1601domainofPTCH1relievesSMOinhibition,leadingtoactivationoftheGLItranscriptionfactorthatregulatesthetranscriptionofSHHtargetgenes(Toftgard,2000).
SurgicalinterventionremainstheprimarytreatmentofchoiceforOKCs.
However,ahighrecurrencerateofupto30%hasbeenfoundfollowingconservativetreatments,suchasenucleationandcurettage,makingitdifficulttodeterminetheoptimalextentofsur‐gicalresection.
AggressivetumorresectioncanleadtotheneedforextensivereconstructivesurgeryandrehabilitationforpatientswithOKCs,causingsignificantmorbidityandnegativelyimpactingtheirqualityoflife(Li,2011;Madras&Lapointe,2008;Miller,Campbell,&Deas,2011;Morgan,Burton,&Qian,2005).
Recently,theSHHpathwaywasidentifiedasapotentialtherapeutictargetwiththein‐troductionofaclinicallyeffectivesmall‐moleculeantagonistoftheSMOreceptor,vismodegib(Rudin,2012).
Indeed,aclinicaltrialinpatientswithGorlinsyndromeshowedthatvismodegibcouldshrinkOKCsinsomepatients,highlightingthepossibilityofprecisionmed‐icineformanagingOKCs(Allyetal.
,2014).
Recently,wereportedfrequentPTCH1(OMIM#601309)muta‐tionsdetectedin16of19(84%)sporadicOKCs(Quetal.
,2015).
Approximately20%ofpatientswithsporadicOKCsdonotappeartocarryaPTCH1mutation;thus,themolecularalterationsofthissubgroupoflesionsremaintobefurtherstudied.
Inthisstudy,wefurtherexpandedthesamplesizetoconfirmPTCH1mutationfrequency,providingthebasisfortargetedtherapeuticstrategy.
Moreover,weappliedwhole‐exomesequencing(WES)foreval‐uationofPTCH1‐negativesporadicOKCsamplesinanattempttodetectnovelgenesinvolvedandbetterunderstandtheoverallmu‐tationlandscape.
2|MATERIALSANDMETHODS2.
1|PatientsandsamplesThirty‐eightpatientswithsporadicOKCswereobtainedfromPekingUniversitySchoolandHospitalofStomatology,nineteenofwhichwerepreviouslydescribedinastudyofPTCH1mutations(Quetal.
,2015).
Informedconsentwasobtained,andallmethodswereperformedinaccordancewiththerelevantguidelinesoftheEthicsCommitteeofthePekingUniversityHealthScienceCenter(No.
2016‐09,DateofApproval:February26,2016).
2.
2|Epitheliumseparation,DNAextraction,andPTCH1sequencingTheepithelialliningswereseparatedfromtheassociatedfibrouscapsule,asdescribedpreviously(Quetal.
,2015).
GenomicDNAwasisolated,andeachofthe22exonscomprisingthePTCH1genewasamplifiedbypolymerasechainreaction(PCR)withspecificprimersandconditions,asdescribedinourpreviousreport(Quetal.
,2015).
SangersequencingwasperformedonanABIPRISM3730GeneticAnalyzer(AppliedBiosystems).
2.
3|LOHassayEightcystswithoutPTCH1mutationwerescreenedforLOH.
Fivemicrosatellitemarkers(D9S253,D9S197,D9S196,D9S287,andPTCH1intra,exon1a)wereexamined.
PCRwasperformedusingafluorescentforwardprimer.
PrimersequenceswereobtainedfromtheNationalCenterforBiotechnologyInformationUniSTSdatabase.
GenomicinstabilitywasdetectedwithanABIPRISM3730GeneticAnalyzer.
Agiveninformativemarkerwasconsid‐eredtodisplayLOHwhena≥2‐folddifferencewasobservedintherelativealleleheightratiobetweenthelesionandmatchedstroma.
CystswithlossofatleasttwolociwereconsideredtoexhibitLOH.
2.
4|Whole‐exomesequencing(WES)andbioinformaticsanalysisFivecystswithoutPTCH1mutationsorLOHsatisfiedWESrequire‐ments.
ThesamplewaspreparedusingAgilentSureSelectHumanAllExon(50M)Kit(AgilentTechnologies)byfollowingthemanufacturerguide.
DNAsequencingwasperformedonanIlluminaHiSeq2500instrument(IlluminaInc)usingstandardprotocols.
Sequencingdataweremappedtothehumanreferencege‐nomeusingtheBurrows–WheelerAligner(BWA)(Li&Durbin,2009).
VariantswerecalledwithGenomeAnalysisToolkit(GATK)(McKennaetal.
,2010).
Fordetectionofsomaticsingle‐nucleotidevariants(SNVs)andindels,pairedsequencingdataforeachlesionandthematchednormalsamplewereanalyzedusingMuTect2(Cibulskisetal.
,2013)andwereannotatedwithANNOVAR.
Wefilteredoutallknownpolymorphismsfrom1,000GenomesanddbSNP.
Highlyconfidentvariantsrequirethefollowingcon‐ditions:(a)non‐silentsomaticvariants;(b)genesmutatedinatleasttwoOKCs;(c)minimumdepthof10*andminimumvariantsupportdepthof4*;and(d)somaticSNVspresentatafrequencyof≥10%orindelsatafrequencyof≥20%inthelesions(Yapetal.
,2014).
FunctionalimpactpredictionswereperformedusingPolyPhen‐2(genetics.
bwh.
harvard.
edu/pph2/)andSIFT(sift.
jcvi.
org/www/SIFT_enst_submit.
html).
Inaddition,weusedphenolyzer(http://phenolyzer.
usc.
edu)topredictthedisease‐causinggenesofOKCs(Yang,Robinson,&Wang,2015).
2.
5|SequencevalidationCyst‐specificvariantswereconfirmedbySangersequencing,andprimersequencesareshowninTablesS1–S2.
Throughaliteraturereviewandphenolyzerprediction,wefocusedontwopotentialdis‐ease‐causinggenes(CDONandMAPK1).
TheconfirmedDNAvari‐antsandfulllengthofCDONandMAPK1werefurtherassessedinavalidationsetofother10OKCs,includingthreePTCH1‐negativecystsandsevenPTCH1‐positivesamples.
1602|2.
6|PathwayenrichmentanalysisWeperformedpathwayenrichmentanalysiswithKOBAS(v3.
0)byexaminingthedistributionofthenon‐synonymouslymutatedgenesidentifiedwithinKyotoEncyclopediaofGenesandGenomes(KEGG).
Significantlyalteredpathwaysweredeterminedbymeasur‐ingpvaluescalculatedonthebasisofhypergeometricdistributionwiththeBenjaminicorrection(correctedpvalueCp.
Cys1043ArgMissenseECL63051/FExon15c.
2419dupAp.
Thr807Asnfs*22FrameshiftECL43128/MExon21c.
3451_3468delinsCAp.
Tyr1151Glnfs*35FrameshiftECL63233/MExon18c.
2893_3124dupp.
Val1042Glyfs*28FrameshiftTM833c61/MExon10c.
1441_1447delp.
Val481Glnfs*8FrameshiftTM3aGenemutationnomenclatureisappliedaccordingtotheguidelinesoftheHumanGenomeVariationSociety.
bNucleotideandaminoacidresiduenumberingarebasedonGenbankentryU59464.
1;+1=AofATGCodon.
cThemutationdemonstratedhomozygosity.
FIGURE1PTCH1alterationsidentifiedinthedetachedepitheliallayersoffreshOKCs.
(a)AmissensePTCH1mutation(c.
3127T>C)wasdetectedintheepitheliallayerofpatient29,resultinginaminoacidsubstitutionsinahighlyconserveddomain,asdemonstratedbysequencealignment.
Thevariantwasabsentfromthestromaltissues.
(b)LOHassayforpatient34,showinglossofthreeDNAmarkers(D9S253,D9S196,andPTCH1intra)inthePTCH1locuswhena≥2‐folddifferencewasobservedintherelativealleleheightratiobetweenthematchedstromaandepitheliallining(arrows)[Colourfigurecanbeviewedatwileyonlinelibrary.
com]|16033|RESULTS3.
1|MutationandLOHofPTCH1insporadicOKCsWedetected15PTCH1mutationsinthefreshepitheliumfrom14of19cases(oneOKCsamplecarriedtwosimultaneousmutations;Table1).
Combinedwithourpreviousresearch(inwhichPTCH1mu‐tationswereobservedin16of19sporadiccases)(Quetal.
,2015),atotalof30outof38patientscarriedPTCH1mutations,corre‐spondingtoamutationfrequencyof79%.
Overall,allthemuta‐tionsweredeterminedtobesomaticbecauseoftheirabsenceinthematchingstromalsamples.
Patient29carriedamissensemutation(c.
3127T>C),resultinginachangefromcysteinetoarginineatcodon1043(Figure1a).
MostoftheremainingvariantsidentifiedwereframeshiftmutationsthatwerepredictedtocausetheprematureterminationofthePTCH1protein.
Theremainingeightcysts(patients17–19and34–38)lackedPTCH1alterations.
Overall,twocysts(patients17and34)showedLOHforatleasttwoloci,whichsuggestedthattheymayhavebeencausedbyLOHofPTCH1insteadofmutations(Figure1b).
3.
2|Identificationofcyst‐specificsomaticvariantsinadiscoverycohortOnaverage,5.
13Gbofhigh‐qualitysequencedataweregeneratedpersample.
Overall,98.
3%ofthesequencereadswerealignedtothehumanreferencegenome(hg19/GRCh37).
Inboththelesionandnormalsamples,a57‐foldmeancoveragewasachievedwith95%ofthebasesonaveragecoveredbyatleast10‐fold.
ThesummarystatisticsofWESarepresentedinTableS3.
AsshowninTable2,wedetected125somaticmutationsinCDSsorsplicingsites(TableS4),correspondingtoanaverage0.
48mu‐tationspermegabase(range,0.
22–0.
72;Figure2a),aconsiderablysmallernumbercomparedwithothertypesofmalignanttumors(Bonillaetal.
,2016;Stranskyetal.
,2011).
All125mutationswereconfirmedinonlyasingleindividual,suchthatnorecurrentalterationwasidentifiedinthediscoverycohort.
Accordingtothescreeningofcandidategenes,weshortlisted31highlyconfidentDNAvariants,amongwhich25variantswerepredictedtobefunctionallydamag‐ing(TableS5).
Twenty‐twoof25variantswereverifiedtobehetero‐zygousSNVsbySangersequencing(Table3).
Moreover,phenolyzeridentified10genesthatwerepredictedaspossibledisease‐causinggenesbasedonthe22confirmedvariants(Figure2b).
3.
3|IdentificationofcandidatedrivermutationsinthevalidationcohortThe22novelvariantsandtwofull‐lengthpredicteddisease‐causinggenes(MAPK1andCDON)weresubjectedtoavalidationscreeninasetofother10OKCs,includingthreePTCH1‐negativeandsevenPTCH1‐positivesamples.
Wefailedtofindrecurrentmutationsinthevalidationset,therebyindicatingthatthe22novelvariantswerenotlikelytobehotspotmutations.
Moreover,MAPK1andCDONTABLE2Overviewofsomaticvariantsfoundinthediscoverysetof5sporadicOKCsSampleIDSNV/IndelFilterstepsVariantsconfirmedNotindbSNPor1,000GenomesInCDS/SplicingsiteHighlyconfidentvariantsaAminoacidchangebMissenseFrameshiftSplicesiteNonsenseSynonymousTotal1810968200046301410919412070004111103650269021214110371047227100634877381287721215736766Total43226384331025125312522Abbreviation:CDS,codingsequence.
a(i)depth≥10*,variantsupportdepth≥4*;(ii)SNVrate≥10%orIndelrate≥20%;(iii)non‐silentvariants;(iv)recurrentvariants.
bVariantswith"Possiblydamaging"to"Damaging"predictedfunctionalimpactsontheencodedprotein,aspredictedbyeffectpredictiontoolSIFTandPolyPhen‐2,havebeenreferredtoasvariantswith"Aminoacidchange".
1604|mutationsrepresenteitherraredriversforpathogenesisorpas‐sengermutationsandarethusunlikelytorepresentcommondrivermutationsinOKCs.
3.
4|AlteredpathwayinsporadicOKCsPathwayanalysesrevealedseveralsignificantalteredpathways,in‐cluding"Basalcellcarcinoma"pathway(TableS6).
Hedgehogsignal‐ingwasavitalcomponentin"Basalcellcarcinoma"pathway,whichsupportedtheessentialroleofSHHsignalinginpathogenesisofOKCs.
4|DISCUSSIONThepresentstudy,togetherwithourpreviousdata(Quetal.
,2015),confirmedthehighPTCH1mutationrate(approximately80%)insporadicOKCs.
GiventhekeyroleofPTCH1inregulatingtheSHHsignalingpathway,theseresultsprovideevidencethatSHHsignalingplaysacriticalroleinOKCdevelopment.
Importantly,theSHHpath‐wayinhibitorvismodegibhasbeenusedasanadjunctivetherapyinpatientswithGorlinsyndrometoreduceOKCsizeanddefinethemarginsneededforsurgicalresection(Allyetal.
,2014).
Thus,sinceapproximately80%ofthesporadicOKCsanalyzedinthesestud‐iesharborPTCH1mutations,ourresultsindicatedthatthisinhibitorcouldalsobeeffectivefortreatingasignificantnumberofsporadicOKCs.
Wecompiledalistof56PTCH1mutationsobservedin44outof90patientswithsporadicOKCsbyourgroup(TableS7)andhaveincludedanadditional15mutationsthatwereidentifiedinthisstudyfrom14of19cases.
Consistentwithpreviousstudies,mostofthemutationsidentified(58/71;82%)arepredictedtoresultintheexpressionoftruncatedPTCH1protein(Figure3).
Asignificantlyhigherfrequency(32/71;45%)clusteredinthetwolargeextra‐cellularloops,wherehedgehogligandbindingoccurs(Lindstrom,Shimokawa,Toftgard,&Zaphiropoulos,2006).
Another"hot"FIGURE2NumberofmutationsincasesbyWESandpredictionofdisease‐causinggenes.
(a)SomaticmutationfrequenciesforfivePTCH1‐negativeOKCs.
(b)TenpredictedgenesinPTCH1‐negativeOKCslistedbyphenolyzerandrankedaccordingtothescore,withahigherscoreindicatingagreaterchancetobeadisease‐causinggene.
Twofull‐lengthgenesvalidatedin10tumors(CDONandMAPK1)aremarked[Colourfigurecanbeviewedatwileyonlinelibrary.
com]|1605mutationregionwasthehighlyconservedsterol‐sensingdomain(SSD)(20/71;28%),whichharborstransmembranedomains2–6(Lindstrometal.
,2006).
However,nomutationswerefoundinthelargeintracellularloop,whichmediatesthenon‐canonicalhedgehogpathway(Yu,Hong,Qu,Chen,&Li,2014).
Afteracarefulandcom‐prehensiveanalysis,neitherPTCH1mutationhotspotsnorapparentgenotype–phenotypecorrelationscouldbeestablished.
WeperformedWESinfivecystsandobtained125somaticvari‐ants.
Althoughwefailedtofindthesamegenemutatedinmorethanonecyst,22novelSNVswereconfirmed.
CDON,amembrane‐asso‐ciatedSHH‐bindingprotein(Tenzenetal.
,2006;Yao,Lum,&Beachy,2006),servesasaco‐receptorwithPTCH1topromoteSHHpath‐wayactivity(Izzietal.
,2011).
MAPK1(extracellularsignal‐regulatedkinase[ERK])isdownstreamofMEK1intheERKpathwayandisassociatedwithcellproliferation.
ActivationoftheERKpathway,asaconsequenceofSHH‐inducedgeneexpression,occursfrequentlyinhumanBCC.
Thus,mutationsinCDONandMAPK1maydirectlyactivateSHHandERKsignalingpathway,respectively,andcontrib‐utetoOKCformation.
The22variantsandtwofull‐lengthgenesCDONandMAPK1weresequencedinavalidationcohortconsistingofthreePTCH1‐negativeandsevenPTCH1‐positivesamples.
Wefailedtofindanymutationsamongthesesamples.
Recently,FrancaetalreportedsomeothermutationsexceptPTCH1in18OKCsbynext‐generationsequencing2,856cancerhotspotmutationsin50oncogenesandtumorsuppres‐sorgenes(Francaetal.
,2018).
Similartoourfinding,thesemutationsTABLE3Detailsof22somaticvariantsconfirmedinPTCH1‐negativeKCOTsGenePatientsDescriptionTranscriptNucleotidechangeAminoaciddefinitionFunctioneffectANK218Ankyrin2,neuronalNM_001148c.
9271G>Tp.
E3091*NonsenseSTAMBPL118STAMbindingprotein‐like1NM_020799c.
503G>Tp.
R168LMissensePIP5K1A18Phosphatidylinositol‐4‐phos‐phate5‐kinase,typeI,alphaNM_001135637c.
1146G>Tp.
R382SMissenseGRM618Glutamatemetabotropicreceptor6NM_000843c.
2424C>Gp.
Q808HMissenseDDX2718DEAD(Asp‐Glu‐Ala‐Asp)boxpolypeptide27NM_017895c.
1865A>Tp.
E622VMissenseSRCAP18Snf2‐relatedCREBBPactiva‐torproteinNM_006662c.
7727C>Ap.
S2576*NonsenseZNF72918Zincfingerprotein729NM_001242680c.
3560C>Ap.
P1187HMissenseCDON18Celladhesionassociated,oncogeneregulatedNM_001243597c.
3406C>Ap.
V1136FMissenseTP5318Tumorproteinp53NM_000546c.
772G>Tp.
E258*NonsenseOR5D1337Olfactoryreceptorfamily5subfamilyDmember13NM_001001967c.
746C>Tp.
A249VMissenseGABRG237ProteintyrosinephosphatasetypeIVA,member3NM_198904c.
1363A>Tp.
I455FMissenseDLG237Disks,largehomolog2(Drosophila)NM_001142700c.
1598G>Ap.
S533FMissenseMAPK137Mitogen‐activatedproteinkinase1NM_002745c.
461T>Cp.
N154SMissensePAX737Pairedbox7NM_001135254c.
947C>Ap.
S316YMissenseITSN237Intersectin2NM_019595c.
4489C>Ap.
L1497IMissenseRYR137ryanodinereceptor1NM_000540c.
2689G>Cp.
D897HMissenseDPYSL538Dihydropyrimidinase‐like5NM_001253723c.
349C>Tp.
R117*NonsensePDE2A38Phosphodiesterase2A,cGMP‐stimulatedNM_002599c.
9G>Tp.
Q3HMissensePIK3CG38Phosphatidylinositol‐4,5‐bisphosphate3‐kinase,catalyticsubunitgammaNM_001282426c.
1666G>Tp.
E556*NonsenseSLC16A1238Solutecarrierfamily16,member12NM_213606c.
1183C>Gp.
P395AMissenseEP40038E1A‐bindingproteinp400NM_015409c.
8272delCp.
P2758Hfs*19FrameshiftTET138Tetmethylcytosinedioxyge‐nase1NM_030625c.
1069G>Tp.
E357*Nonsense1606|occurredinonesampleeachandtheyfailedtodetectrecurrenthotspotmutations.
ThemutationsdetectedbyFrancaweredifferentfromthatfoundinourstudysuggestingthatthesecancerhotspotmutationsmightoccurrarelyinOKCs.
Thus,weinferredthatmuta‐tionsoccurringintheSHHpathwayregulatorPTCH1(~80%)likelyplayacentralroleinOKCs,whereasotherraremutations(~20%)scat‐teredinremainingOKCcaseswereindependentoftheSHHpath‐way.
OurfindingsconfirmedthefunctionalinvolvementoftheSHHpathwayinthemajorityofOKCsandhighlightedtheusefulnessoftreatmentwithmolecularlytargeteddrugsthatinhibitthispathway,suchasvismodegib.
ACKNOWLEDGEMENTSWegratefullyacknowledgethepatientswhoparticipatedinthisstudy.
ThisworkwassupportedResearchGrantsfromtheNationalNatureScienceFoundationofChina(81671006and81030018),theBeijingNatureScienceFoundation(7172238),andtheFundamentalResearchFundsfortheCentralUniversities,NankaiUniversity(63191118).
CONFLICTOFINTERESTDeclarednone.
AUTHORCONTRIBUTIONSLiTandChenFdesignedthestudy.
QuJandZhangJcollectedsamples,performedhigh‐throughputsequencingdataanalysisanddraftedthemanuscript.
ZhangHandLiXconductedqualitycontrolofsequencingdata.
HongY,ZhaiJandWangYperformedsampleandhigh‐throughputsequencinglibrarypreparation.
DATAAVAILABILITYSTATEMENTAlldatacouldbeavailableinthisarticleanditsadditionalfiles.
TherawsequencedatareportedinthispaperhavebeendepositedintheGenomeSequenceArchive(Genomics,Proteomics&Bioinformatics2017)(Wangetal.
,2017)inBIGDataCenter(NucleicAcidsRes2018)(BIGDataCenterMembers,2019),BeijingInstituteofGenomics(BIG),ChineseAcademyofSciences,underaccessionnumbersCRA001584,CRA001584,thatarepubliclyaccessibleathttp://bigd.
big.
ac.
cn/gsa.
ORCIDTiejunLihttps://orcid.
org/0000-0001-5983-2985REFERENCESAlly,M.
S.
,Tang,J.
Y.
,Joseph,T.
,Thompson,B.
,Lindgren,J.
,Raphael,M.
A.
,…Epstein,E.
H.
(2014).
Theuseofvismodegibtoshrinkkeratocys‐ticodontogenictumorsinpatientswithbasalcellnevussyndrome.
JAMADermatology,150(5),542–545.
https://doi.
org/10.
1001/jamadermatol.
2013.
7444Bonilla,X.
,Parmentier,L.
,King,B.
,Bezrukov,F.
,Kaya,G.
,Zoete,V.
,…Nikolaev,S.
I.
(2016).
Genomicanalysisidentifiesnewdriversandprogressionpathwaysinskinbasalcellcarcinoma.
NatureGenetics,48(4),398–406.
https://doi.
org/10.
1038/ng.
3525Cibulskis,K.
,Lawrence,M.
S.
,Carter,S.
L.
,Sivachenko,A.
,Jaffe,D.
,Sougnez,C.
,…Getz,G.
(2013).
Sensitivedetectionofsomaticpointmutationsinimpureandheterogeneouscancersamples.
NatureBiotechnology,31(3),213–219.
https://doi.
org/10.
1038/nbt.
2514FIGURE3Distributionpatternof71PTCH1mutationsrelatedtothedifferentdomainsofthepatchedprotein.
ThethicklineindicatestheSSD[Colourfigurecanbeviewedatwileyonlinelibrary.
com]|1607Frana,J.
A.
,deSousa,S.
F.
,Diniz,M.
G.
,Pereira,T.
D.
S.
F.
,deResende,T.
A.
C.
,Santos,J.
N.
D.
,…Gomes,C.
C.
(2018).
AbsenceofBRAFV600Emutationinodontogenickeratocysts.
JournalofOralPathologyandMedicine,47(2),186–191.
https://doi.
org/10.
1111/jop.
12671Izzi,L.
,Lévesque,M.
,Morin,S.
,Laniel,D.
,Wilkes,B.
C.
,Mille,F.
,…Charron,F.
(2011).
BocandGas1eachformdistinctShhreceptorcomplexeswithPtch1andarerequiredforShh‐mediatedcellprolifer‐ation.
DevelopmentalCell,20(6),788–801.
https://doi.
org/10.
1016/j.
devcel.
2011.
04.
017Li,H.
,&Durbin,R.
(2009).
FastandaccurateshortreadalignmentwithBurrows‐Wheelertransform.
Bioinformatics,25(14),1754–1760.
https://doi.
org/10.
1093/bioinformatics/btp324Li,T.
J.
(2011).
Theodontogenickeratocyst:Acyst,oracysticneo‐plasmJournalofDentalResearch,90(2),133–142.
https://doi.
org/10.
1177/0022034510379016Lindstrom,E.
,Shimokawa,T.
,Toftgard,R.
,&Zaphiropoulos,P.
G.
(2006).
PTCHmutations:Distributionandanalyses.
HumanMutation,27(3),215–219.
https://doi.
org/10.
1002/humu.
20296Madras,J.
,&Lapointe,H.
(2008).
Keratocysticodontogenictumour:Reclassificationoftheodontogenickeratocystfromcysttotumour.
Journal/CanadianDentalAssociation,74(2),165–165h.
McKenna,A.
,Hanna,M.
,Banks,E.
,Sivachenko,A.
,Cibulskis,K.
,Kernytsky,A.
,…DePristo,M.
A.
(2010).
TheGenomeAnalysisToolkit:AMapReduceframeworkforanalyzingnext‐generationDNAsequencingdata.
GenomeResearch,20(9),1297–1303.
https://doi.
org/10.
1101/gr.
107524.
110Miller,N.
M.
,Campbell,C.
M.
,&Deas,D.
E.
(2011).
Reconstructivesurgi‐calmanagementofarecurrentodontogenickeratocyst:Casereportand1‐yearfollow‐up.
TheJournaloftheMichiganDentalAssociation,93(9),38–43.
Morgan,T.
A.
,Burton,C.
C.
,&Qian,F.
(2005).
Aretrospectivereviewoftreatmentoftheodontogenickeratocyst.
JournalofOralandMaxillofacialSurgery,63(5),635–639.
https://doi.
org/10.
1016/j.
joms.
2004.
07.
026Partridge,M.
,&Towers,J.
F.
(1987).
Theprimordialcyst(odontogenickeratocyst):Itstumour‐likecharacteristicsandbehaviour.
TheBritishJournalofOral&MaxillofacialSurgery,25(4),271–279.
Qu,J.
,Yu,F.
,Hong,Y.
,Guo,Y.
,Sun,L.
,Li,X.
,…Li,T.
(2015).
UnderestimatedPTCH1mutationrateinsporadickeratocysticodontogenictumors.
OralOncology,51(1),40–45.
https://doi.
org/10.
1016/j.
oraloncology.
2014.
09.
016Rudin,C.
M.
(2012).
Vismodegib.
ClinicalCancerResearch:AnOfficialJournaloftheAmericanAssociationforCancerResearch,18(12),3218–3222.
https://doi.
org/10.
1158/1078-0432.
CCR-12-0568Stone,D.
M.
,Hynes,M.
,Armanini,M.
,Swanson,T.
A.
,Gu,Q.
,Johnson,R.
L.
,…Rosenthal,A.
(1996).
Thetumour‐suppressorgenepatcheden‐codesacandidatereceptorforSonichedgehog.
Nature,384(6605),129–134.
https://doi.
org/10.
1038/384129a0Stransky,N.
,Egloff,A.
M.
,Tward,A.
D.
,Kostic,A.
D.
,Cibulskis,K.
,Sivachenko,A.
,…Grandis,J.
R.
(2011).
Themutationallandscapeofheadandnecksquamouscellcarcinoma.
Science,333(6046),1157–1160.
https://doi.
org/10.
1126/science.
1208130Tenzen,T.
,Allen,B.
L.
,Cole,F.
,Kang,J.
S.
,Krauss,R.
S.
,&McMahon,A.
P.
(2006).
ThecellsurfacemembraneproteinsCdoandBocarecomponentsandtargetsoftheHedgehogsignalingpathwayandfeedbacknetworkinmice.
DevelopmentalCell,10(5),647–656.
https://doi.
org/10.
1016/j.
devcel.
2006.
04.
004Toftgard,R.
(2000).
Hedgehogsignallingincancer.
CellularandMolecularLifeSciences,57(12),1720–1731.
https://doi.
org/10.
1007/PL00000654Wang,Y.
,Song,F.
,Zhu,J.
,Zhang,S.
,Yang,Y.
,Chen,T.
,…Zhao,W.
(2017).
GSA:Genomesequencearchive*.
GenomicsProteomicsBioinformatics,15(1),14–18.
https://doi.
org/10.
1016/j.
gpb.
2017.
01.
001Yang,H.
,Robinson,P.
N.
,&Wang,K.
(2015).
Phenolyzer:Phenotype‐basedprioritizationofcandidategenesforhumandiseases.
NatureMethods,12(9),841–843.
https://doi.
org/10.
1038/NMETH.
3484Yao,S.
,Lum,L.
,&Beachy,P.
(2006).
Theihogcell‐surfaceproteinsbindHedgehogandmediatepathwayactivation.
Cell,125(2),343–357.
https://doi.
org/10.
1016/j.
cell.
2006.
02.
040Yap,K.
L.
,Kiyotani,K.
,Tamura,K.
,Antic,T.
,Jang,M.
,Montoya,M.
,…Nakamura,Y.
(2014).
Whole‐exomesequencingofmuscle‐invasivebladdercanceridentifiesrecurrentmutationsofUNC5Candprog‐nosticimportanceofDNArepairgenemutationsonsurvival.
ClinicalCancerResearch:AnOfficialJournaloftheAmericanAssociationforCancerResearch,20(24),6605–6617.
https://doi.
org/10.
1158/1078-0432.
CCR-14-0257Yu,F.
Y.
,Hong,Y.
Y.
,Qu,J.
F.
,Chen,F.
,&Li,T.
J.
(2014).
Thelargein‐tracellularloopofptch1mediatesthenon‐canonicalHedgehogpathwaythroughcyclinB1innevoidbasalcellcarcinomasyndrome.
InternationalJournalofMolecularMedicine,34(2),507–512.
https://doi.
org/10.
3892/ijmm.
2014.
1783BIGDataCenterMembers(2019).
DatabaseresourcesoftheBIGDataCenterin2019.
NucleicAcidsResearch,47(D1),D8–D14.
https://doi.
org/10.
1093/nar/gky993SUPPORTINGINFORMATIONAdditionalsupportinginformationmaybefoundonlineintheSupportingInformationsectionattheendofthearticle.
Howtocitethisarticle:QuJ,ZhangJ,ZhangH,etal.
PTCH1alterationsarefrequentbutothergeneticalterationsarerareinsporadicodontogenickeratocysts.
OralDis.
2019;25:1600–1607.
https://doi.
org/10.
1111/odi.
13135
2019;25:1600–1607.
wileyonlinelibrary.
com/journal/odi1|INTRODUCTIONOdontogenickeratocysts(OKCs,alsoknownaskeratocysticodon‐togenictumors)arelocallyaggressivecysticlesionsofthejawthatarethroughtoarisefromtheodontogenicepithelium(Partridge&Towers,1987).
GorlinsyndromeisanautosomaldominantdisorderpredisposingaffectedindividualstothedevelopmentofmultipleOKCs,andstudiesonpatientswithGorlinsyndromehaveinitiallyshownapivotalroleforthesonichedgehog(SHH)pathwayinOKCdevelopment.
FurtherresearchhasconfirmedthecentralroleoftheSHH/patched1(PTCH1)/smoothened(SMO)pathwayinsporadicOKCs.
PTCH1normallyactstoblocktheactivityofSMO(Stoneetal.
,1996).
Inparticular,thebindingofSHHtotheextracellularReceived:5April2019|Revised:15May2019|Accepted:28May2019DOI:10.
1111/odi.
13135ORIGINALARTICLEPTCH1alterationsarefrequentbutothergeneticalterationsarerareinsporadicodontogenickeratocystsJiafeiQu1,2|JianyunZhang1|HeyuZhang3|XuefenLi3|YingyingHong1|JiemeiZhai4|YanjinWang1|FengChen3|TiejunLi12019JohnWiley&SonsA/S.
PublishedbyJohnWiley&SonsLtd.
AllrightsreservedJiafeiQuandJianyunZhangauthorscontributedequallyandsharefirstauthorship.
1DepartmentofOralPathology,PekingUniversitySchoolandHospitalofStomatology,Beijing,China2InternationalVIPDentalClinic,TianjinStomatologicalHospital,StomatologicalHospitalofNankaiUniversity,Tianjin,China3CentralLaboratory,PekingUniversitySchoolandHospitalofStomatology,Beijing,China4DepartmentofOralPathology,SchoolofStomatology,KunmingMedicalUniversity,Kunming,ChinaCorrespondenceTiejunLi,DepartmentofOralPathology,PekingUniversitySchoolandHospitalofStomatology,22SouthZhongguancunAvenue,HaidianDistrict,Beijing100081,China.
Email:litiejun22@vip.
sina.
comFengChen,CentralLaboratory,PekingUniversitySchoolandHospitalofStomatology,22SouthZhongguancunAvenue,HaidianDistrict,Beijing100081,China.
Email:chenfeng2011@hsc.
pku.
edu.
cnFundinginformationtheBeijingNatureScienceFoundation,Grant/AwardNumber:7172238;theNationalNatureScienceFoundationofChina,Grant/AwardNumber:81030018and81671006;theFundamentalResearchFundsfortheCentralUniversities,NankaiUniversity,Grant/AwardNumber:63191118AbstractObjective:Odontogenickeratocysts(OKCs)arebenignjawlesionswithhighgrowthpotentialandpropensityforrecurrence.
OurpreviousstudyrevealedthatPTCH1mu‐tations,whichwerefrequentlydetectedinsporadicOKCs,mightbeunderestimatedduetothemaskingeffectofthestromalcomponentswithinthetestedtissues.
WeaimedtoconfirmtheseresultsinlargerscaleandfurtherpresenttheunbiasedviewofthegenomicbasisofsporadicOKCsexceptPTCH1.
Materialsandmethods:WeanalyzedPTCH1mutationsinadditional19samples.
Usingwhole‐exomesequencing(WES),wefurthercharacterizedthemutationallandscapeoffivesporadicOKCsampleslackingPTCH1mutationandlossofhet‐erozygosity(LOH).
Results:Combinedwithourpreviouslyreported19cases,thirtyof38(79%)casesharboredPTCH1mutations.
Throughwhole‐exomesequencingandintegrativeanal‐ysis,22novelmutationswereconfirmedamongfivePTCH1‐negativesamples.
NorecurrentmutationswereidentifiedintheWESsamplesandvalidationcohortof10OKCs.
Conclusions:OurdatafurtherconfirmedthefrequentPTCH1mutationandotherraregeneticalterationsinsporadicOKCs,highlightingthecentralroleofSHHsignal‐ingpathway.
InPTCH1‐negativecases,otherraremutationsscatteredinasubsetofOKCswereindependentoftheSHHpathway.
TheseresultssuggestedthatanSHHinhibitormaybeeffectivetotreatthemajorityofOKCs.
KEYWORDSPTCH1mutation,odontogenickeratocysts,whole‐exomesequencing|1601domainofPTCH1relievesSMOinhibition,leadingtoactivationoftheGLItranscriptionfactorthatregulatesthetranscriptionofSHHtargetgenes(Toftgard,2000).
SurgicalinterventionremainstheprimarytreatmentofchoiceforOKCs.
However,ahighrecurrencerateofupto30%hasbeenfoundfollowingconservativetreatments,suchasenucleationandcurettage,makingitdifficulttodeterminetheoptimalextentofsur‐gicalresection.
AggressivetumorresectioncanleadtotheneedforextensivereconstructivesurgeryandrehabilitationforpatientswithOKCs,causingsignificantmorbidityandnegativelyimpactingtheirqualityoflife(Li,2011;Madras&Lapointe,2008;Miller,Campbell,&Deas,2011;Morgan,Burton,&Qian,2005).
Recently,theSHHpathwaywasidentifiedasapotentialtherapeutictargetwiththein‐troductionofaclinicallyeffectivesmall‐moleculeantagonistoftheSMOreceptor,vismodegib(Rudin,2012).
Indeed,aclinicaltrialinpatientswithGorlinsyndromeshowedthatvismodegibcouldshrinkOKCsinsomepatients,highlightingthepossibilityofprecisionmed‐icineformanagingOKCs(Allyetal.
,2014).
Recently,wereportedfrequentPTCH1(OMIM#601309)muta‐tionsdetectedin16of19(84%)sporadicOKCs(Quetal.
,2015).
Approximately20%ofpatientswithsporadicOKCsdonotappeartocarryaPTCH1mutation;thus,themolecularalterationsofthissubgroupoflesionsremaintobefurtherstudied.
Inthisstudy,wefurtherexpandedthesamplesizetoconfirmPTCH1mutationfrequency,providingthebasisfortargetedtherapeuticstrategy.
Moreover,weappliedwhole‐exomesequencing(WES)foreval‐uationofPTCH1‐negativesporadicOKCsamplesinanattempttodetectnovelgenesinvolvedandbetterunderstandtheoverallmu‐tationlandscape.
2|MATERIALSANDMETHODS2.
1|PatientsandsamplesThirty‐eightpatientswithsporadicOKCswereobtainedfromPekingUniversitySchoolandHospitalofStomatology,nineteenofwhichwerepreviouslydescribedinastudyofPTCH1mutations(Quetal.
,2015).
Informedconsentwasobtained,andallmethodswereperformedinaccordancewiththerelevantguidelinesoftheEthicsCommitteeofthePekingUniversityHealthScienceCenter(No.
2016‐09,DateofApproval:February26,2016).
2.
2|Epitheliumseparation,DNAextraction,andPTCH1sequencingTheepithelialliningswereseparatedfromtheassociatedfibrouscapsule,asdescribedpreviously(Quetal.
,2015).
GenomicDNAwasisolated,andeachofthe22exonscomprisingthePTCH1genewasamplifiedbypolymerasechainreaction(PCR)withspecificprimersandconditions,asdescribedinourpreviousreport(Quetal.
,2015).
SangersequencingwasperformedonanABIPRISM3730GeneticAnalyzer(AppliedBiosystems).
2.
3|LOHassayEightcystswithoutPTCH1mutationwerescreenedforLOH.
Fivemicrosatellitemarkers(D9S253,D9S197,D9S196,D9S287,andPTCH1intra,exon1a)wereexamined.
PCRwasperformedusingafluorescentforwardprimer.
PrimersequenceswereobtainedfromtheNationalCenterforBiotechnologyInformationUniSTSdatabase.
GenomicinstabilitywasdetectedwithanABIPRISM3730GeneticAnalyzer.
Agiveninformativemarkerwasconsid‐eredtodisplayLOHwhena≥2‐folddifferencewasobservedintherelativealleleheightratiobetweenthelesionandmatchedstroma.
CystswithlossofatleasttwolociwereconsideredtoexhibitLOH.
2.
4|Whole‐exomesequencing(WES)andbioinformaticsanalysisFivecystswithoutPTCH1mutationsorLOHsatisfiedWESrequire‐ments.
ThesamplewaspreparedusingAgilentSureSelectHumanAllExon(50M)Kit(AgilentTechnologies)byfollowingthemanufacturerguide.
DNAsequencingwasperformedonanIlluminaHiSeq2500instrument(IlluminaInc)usingstandardprotocols.
Sequencingdataweremappedtothehumanreferencege‐nomeusingtheBurrows–WheelerAligner(BWA)(Li&Durbin,2009).
VariantswerecalledwithGenomeAnalysisToolkit(GATK)(McKennaetal.
,2010).
Fordetectionofsomaticsingle‐nucleotidevariants(SNVs)andindels,pairedsequencingdataforeachlesionandthematchednormalsamplewereanalyzedusingMuTect2(Cibulskisetal.
,2013)andwereannotatedwithANNOVAR.
Wefilteredoutallknownpolymorphismsfrom1,000GenomesanddbSNP.
Highlyconfidentvariantsrequirethefollowingcon‐ditions:(a)non‐silentsomaticvariants;(b)genesmutatedinatleasttwoOKCs;(c)minimumdepthof10*andminimumvariantsupportdepthof4*;and(d)somaticSNVspresentatafrequencyof≥10%orindelsatafrequencyof≥20%inthelesions(Yapetal.
,2014).
FunctionalimpactpredictionswereperformedusingPolyPhen‐2(genetics.
bwh.
harvard.
edu/pph2/)andSIFT(sift.
jcvi.
org/www/SIFT_enst_submit.
html).
Inaddition,weusedphenolyzer(http://phenolyzer.
usc.
edu)topredictthedisease‐causinggenesofOKCs(Yang,Robinson,&Wang,2015).
2.
5|SequencevalidationCyst‐specificvariantswereconfirmedbySangersequencing,andprimersequencesareshowninTablesS1–S2.
Throughaliteraturereviewandphenolyzerprediction,wefocusedontwopotentialdis‐ease‐causinggenes(CDONandMAPK1).
TheconfirmedDNAvari‐antsandfulllengthofCDONandMAPK1werefurtherassessedinavalidationsetofother10OKCs,includingthreePTCH1‐negativecystsandsevenPTCH1‐positivesamples.
1602|2.
6|PathwayenrichmentanalysisWeperformedpathwayenrichmentanalysiswithKOBAS(v3.
0)byexaminingthedistributionofthenon‐synonymouslymutatedgenesidentifiedwithinKyotoEncyclopediaofGenesandGenomes(KEGG).
Significantlyalteredpathwaysweredeterminedbymeasur‐ingpvaluescalculatedonthebasisofhypergeometricdistributionwiththeBenjaminicorrection(correctedpvalueCp.
Cys1043ArgMissenseECL63051/FExon15c.
2419dupAp.
Thr807Asnfs*22FrameshiftECL43128/MExon21c.
3451_3468delinsCAp.
Tyr1151Glnfs*35FrameshiftECL63233/MExon18c.
2893_3124dupp.
Val1042Glyfs*28FrameshiftTM833c61/MExon10c.
1441_1447delp.
Val481Glnfs*8FrameshiftTM3aGenemutationnomenclatureisappliedaccordingtotheguidelinesoftheHumanGenomeVariationSociety.
bNucleotideandaminoacidresiduenumberingarebasedonGenbankentryU59464.
1;+1=AofATGCodon.
cThemutationdemonstratedhomozygosity.
FIGURE1PTCH1alterationsidentifiedinthedetachedepitheliallayersoffreshOKCs.
(a)AmissensePTCH1mutation(c.
3127T>C)wasdetectedintheepitheliallayerofpatient29,resultinginaminoacidsubstitutionsinahighlyconserveddomain,asdemonstratedbysequencealignment.
Thevariantwasabsentfromthestromaltissues.
(b)LOHassayforpatient34,showinglossofthreeDNAmarkers(D9S253,D9S196,andPTCH1intra)inthePTCH1locuswhena≥2‐folddifferencewasobservedintherelativealleleheightratiobetweenthematchedstromaandepitheliallining(arrows)[Colourfigurecanbeviewedatwileyonlinelibrary.
com]|16033|RESULTS3.
1|MutationandLOHofPTCH1insporadicOKCsWedetected15PTCH1mutationsinthefreshepitheliumfrom14of19cases(oneOKCsamplecarriedtwosimultaneousmutations;Table1).
Combinedwithourpreviousresearch(inwhichPTCH1mu‐tationswereobservedin16of19sporadiccases)(Quetal.
,2015),atotalof30outof38patientscarriedPTCH1mutations,corre‐spondingtoamutationfrequencyof79%.
Overall,allthemuta‐tionsweredeterminedtobesomaticbecauseoftheirabsenceinthematchingstromalsamples.
Patient29carriedamissensemutation(c.
3127T>C),resultinginachangefromcysteinetoarginineatcodon1043(Figure1a).
MostoftheremainingvariantsidentifiedwereframeshiftmutationsthatwerepredictedtocausetheprematureterminationofthePTCH1protein.
Theremainingeightcysts(patients17–19and34–38)lackedPTCH1alterations.
Overall,twocysts(patients17and34)showedLOHforatleasttwoloci,whichsuggestedthattheymayhavebeencausedbyLOHofPTCH1insteadofmutations(Figure1b).
3.
2|Identificationofcyst‐specificsomaticvariantsinadiscoverycohortOnaverage,5.
13Gbofhigh‐qualitysequencedataweregeneratedpersample.
Overall,98.
3%ofthesequencereadswerealignedtothehumanreferencegenome(hg19/GRCh37).
Inboththelesionandnormalsamples,a57‐foldmeancoveragewasachievedwith95%ofthebasesonaveragecoveredbyatleast10‐fold.
ThesummarystatisticsofWESarepresentedinTableS3.
AsshowninTable2,wedetected125somaticmutationsinCDSsorsplicingsites(TableS4),correspondingtoanaverage0.
48mu‐tationspermegabase(range,0.
22–0.
72;Figure2a),aconsiderablysmallernumbercomparedwithothertypesofmalignanttumors(Bonillaetal.
,2016;Stranskyetal.
,2011).
All125mutationswereconfirmedinonlyasingleindividual,suchthatnorecurrentalterationwasidentifiedinthediscoverycohort.
Accordingtothescreeningofcandidategenes,weshortlisted31highlyconfidentDNAvariants,amongwhich25variantswerepredictedtobefunctionallydamag‐ing(TableS5).
Twenty‐twoof25variantswereverifiedtobehetero‐zygousSNVsbySangersequencing(Table3).
Moreover,phenolyzeridentified10genesthatwerepredictedaspossibledisease‐causinggenesbasedonthe22confirmedvariants(Figure2b).
3.
3|IdentificationofcandidatedrivermutationsinthevalidationcohortThe22novelvariantsandtwofull‐lengthpredicteddisease‐causinggenes(MAPK1andCDON)weresubjectedtoavalidationscreeninasetofother10OKCs,includingthreePTCH1‐negativeandsevenPTCH1‐positivesamples.
Wefailedtofindrecurrentmutationsinthevalidationset,therebyindicatingthatthe22novelvariantswerenotlikelytobehotspotmutations.
Moreover,MAPK1andCDONTABLE2Overviewofsomaticvariantsfoundinthediscoverysetof5sporadicOKCsSampleIDSNV/IndelFilterstepsVariantsconfirmedNotindbSNPor1,000GenomesInCDS/SplicingsiteHighlyconfidentvariantsaAminoacidchangebMissenseFrameshiftSplicesiteNonsenseSynonymousTotal1810968200046301410919412070004111103650269021214110371047227100634877381287721215736766Total43226384331025125312522Abbreviation:CDS,codingsequence.
a(i)depth≥10*,variantsupportdepth≥4*;(ii)SNVrate≥10%orIndelrate≥20%;(iii)non‐silentvariants;(iv)recurrentvariants.
bVariantswith"Possiblydamaging"to"Damaging"predictedfunctionalimpactsontheencodedprotein,aspredictedbyeffectpredictiontoolSIFTandPolyPhen‐2,havebeenreferredtoasvariantswith"Aminoacidchange".
1604|mutationsrepresenteitherraredriversforpathogenesisorpas‐sengermutationsandarethusunlikelytorepresentcommondrivermutationsinOKCs.
3.
4|AlteredpathwayinsporadicOKCsPathwayanalysesrevealedseveralsignificantalteredpathways,in‐cluding"Basalcellcarcinoma"pathway(TableS6).
Hedgehogsignal‐ingwasavitalcomponentin"Basalcellcarcinoma"pathway,whichsupportedtheessentialroleofSHHsignalinginpathogenesisofOKCs.
4|DISCUSSIONThepresentstudy,togetherwithourpreviousdata(Quetal.
,2015),confirmedthehighPTCH1mutationrate(approximately80%)insporadicOKCs.
GiventhekeyroleofPTCH1inregulatingtheSHHsignalingpathway,theseresultsprovideevidencethatSHHsignalingplaysacriticalroleinOKCdevelopment.
Importantly,theSHHpath‐wayinhibitorvismodegibhasbeenusedasanadjunctivetherapyinpatientswithGorlinsyndrometoreduceOKCsizeanddefinethemarginsneededforsurgicalresection(Allyetal.
,2014).
Thus,sinceapproximately80%ofthesporadicOKCsanalyzedinthesestud‐iesharborPTCH1mutations,ourresultsindicatedthatthisinhibitorcouldalsobeeffectivefortreatingasignificantnumberofsporadicOKCs.
Wecompiledalistof56PTCH1mutationsobservedin44outof90patientswithsporadicOKCsbyourgroup(TableS7)andhaveincludedanadditional15mutationsthatwereidentifiedinthisstudyfrom14of19cases.
Consistentwithpreviousstudies,mostofthemutationsidentified(58/71;82%)arepredictedtoresultintheexpressionoftruncatedPTCH1protein(Figure3).
Asignificantlyhigherfrequency(32/71;45%)clusteredinthetwolargeextra‐cellularloops,wherehedgehogligandbindingoccurs(Lindstrom,Shimokawa,Toftgard,&Zaphiropoulos,2006).
Another"hot"FIGURE2NumberofmutationsincasesbyWESandpredictionofdisease‐causinggenes.
(a)SomaticmutationfrequenciesforfivePTCH1‐negativeOKCs.
(b)TenpredictedgenesinPTCH1‐negativeOKCslistedbyphenolyzerandrankedaccordingtothescore,withahigherscoreindicatingagreaterchancetobeadisease‐causinggene.
Twofull‐lengthgenesvalidatedin10tumors(CDONandMAPK1)aremarked[Colourfigurecanbeviewedatwileyonlinelibrary.
com]|1605mutationregionwasthehighlyconservedsterol‐sensingdomain(SSD)(20/71;28%),whichharborstransmembranedomains2–6(Lindstrometal.
,2006).
However,nomutationswerefoundinthelargeintracellularloop,whichmediatesthenon‐canonicalhedgehogpathway(Yu,Hong,Qu,Chen,&Li,2014).
Afteracarefulandcom‐prehensiveanalysis,neitherPTCH1mutationhotspotsnorapparentgenotype–phenotypecorrelationscouldbeestablished.
WeperformedWESinfivecystsandobtained125somaticvari‐ants.
Althoughwefailedtofindthesamegenemutatedinmorethanonecyst,22novelSNVswereconfirmed.
CDON,amembrane‐asso‐ciatedSHH‐bindingprotein(Tenzenetal.
,2006;Yao,Lum,&Beachy,2006),servesasaco‐receptorwithPTCH1topromoteSHHpath‐wayactivity(Izzietal.
,2011).
MAPK1(extracellularsignal‐regulatedkinase[ERK])isdownstreamofMEK1intheERKpathwayandisassociatedwithcellproliferation.
ActivationoftheERKpathway,asaconsequenceofSHH‐inducedgeneexpression,occursfrequentlyinhumanBCC.
Thus,mutationsinCDONandMAPK1maydirectlyactivateSHHandERKsignalingpathway,respectively,andcontrib‐utetoOKCformation.
The22variantsandtwofull‐lengthgenesCDONandMAPK1weresequencedinavalidationcohortconsistingofthreePTCH1‐negativeandsevenPTCH1‐positivesamples.
Wefailedtofindanymutationsamongthesesamples.
Recently,FrancaetalreportedsomeothermutationsexceptPTCH1in18OKCsbynext‐generationsequencing2,856cancerhotspotmutationsin50oncogenesandtumorsuppres‐sorgenes(Francaetal.
,2018).
Similartoourfinding,thesemutationsTABLE3Detailsof22somaticvariantsconfirmedinPTCH1‐negativeKCOTsGenePatientsDescriptionTranscriptNucleotidechangeAminoaciddefinitionFunctioneffectANK218Ankyrin2,neuronalNM_001148c.
9271G>Tp.
E3091*NonsenseSTAMBPL118STAMbindingprotein‐like1NM_020799c.
503G>Tp.
R168LMissensePIP5K1A18Phosphatidylinositol‐4‐phos‐phate5‐kinase,typeI,alphaNM_001135637c.
1146G>Tp.
R382SMissenseGRM618Glutamatemetabotropicreceptor6NM_000843c.
2424C>Gp.
Q808HMissenseDDX2718DEAD(Asp‐Glu‐Ala‐Asp)boxpolypeptide27NM_017895c.
1865A>Tp.
E622VMissenseSRCAP18Snf2‐relatedCREBBPactiva‐torproteinNM_006662c.
7727C>Ap.
S2576*NonsenseZNF72918Zincfingerprotein729NM_001242680c.
3560C>Ap.
P1187HMissenseCDON18Celladhesionassociated,oncogeneregulatedNM_001243597c.
3406C>Ap.
V1136FMissenseTP5318Tumorproteinp53NM_000546c.
772G>Tp.
E258*NonsenseOR5D1337Olfactoryreceptorfamily5subfamilyDmember13NM_001001967c.
746C>Tp.
A249VMissenseGABRG237ProteintyrosinephosphatasetypeIVA,member3NM_198904c.
1363A>Tp.
I455FMissenseDLG237Disks,largehomolog2(Drosophila)NM_001142700c.
1598G>Ap.
S533FMissenseMAPK137Mitogen‐activatedproteinkinase1NM_002745c.
461T>Cp.
N154SMissensePAX737Pairedbox7NM_001135254c.
947C>Ap.
S316YMissenseITSN237Intersectin2NM_019595c.
4489C>Ap.
L1497IMissenseRYR137ryanodinereceptor1NM_000540c.
2689G>Cp.
D897HMissenseDPYSL538Dihydropyrimidinase‐like5NM_001253723c.
349C>Tp.
R117*NonsensePDE2A38Phosphodiesterase2A,cGMP‐stimulatedNM_002599c.
9G>Tp.
Q3HMissensePIK3CG38Phosphatidylinositol‐4,5‐bisphosphate3‐kinase,catalyticsubunitgammaNM_001282426c.
1666G>Tp.
E556*NonsenseSLC16A1238Solutecarrierfamily16,member12NM_213606c.
1183C>Gp.
P395AMissenseEP40038E1A‐bindingproteinp400NM_015409c.
8272delCp.
P2758Hfs*19FrameshiftTET138Tetmethylcytosinedioxyge‐nase1NM_030625c.
1069G>Tp.
E357*Nonsense1606|occurredinonesampleeachandtheyfailedtodetectrecurrenthotspotmutations.
ThemutationsdetectedbyFrancaweredifferentfromthatfoundinourstudysuggestingthatthesecancerhotspotmutationsmightoccurrarelyinOKCs.
Thus,weinferredthatmuta‐tionsoccurringintheSHHpathwayregulatorPTCH1(~80%)likelyplayacentralroleinOKCs,whereasotherraremutations(~20%)scat‐teredinremainingOKCcaseswereindependentoftheSHHpath‐way.
OurfindingsconfirmedthefunctionalinvolvementoftheSHHpathwayinthemajorityofOKCsandhighlightedtheusefulnessoftreatmentwithmolecularlytargeteddrugsthatinhibitthispathway,suchasvismodegib.
ACKNOWLEDGEMENTSWegratefullyacknowledgethepatientswhoparticipatedinthisstudy.
ThisworkwassupportedResearchGrantsfromtheNationalNatureScienceFoundationofChina(81671006and81030018),theBeijingNatureScienceFoundation(7172238),andtheFundamentalResearchFundsfortheCentralUniversities,NankaiUniversity(63191118).
CONFLICTOFINTERESTDeclarednone.
AUTHORCONTRIBUTIONSLiTandChenFdesignedthestudy.
QuJandZhangJcollectedsamples,performedhigh‐throughputsequencingdataanalysisanddraftedthemanuscript.
ZhangHandLiXconductedqualitycontrolofsequencingdata.
HongY,ZhaiJandWangYperformedsampleandhigh‐throughputsequencinglibrarypreparation.
DATAAVAILABILITYSTATEMENTAlldatacouldbeavailableinthisarticleanditsadditionalfiles.
TherawsequencedatareportedinthispaperhavebeendepositedintheGenomeSequenceArchive(Genomics,Proteomics&Bioinformatics2017)(Wangetal.
,2017)inBIGDataCenter(NucleicAcidsRes2018)(BIGDataCenterMembers,2019),BeijingInstituteofGenomics(BIG),ChineseAcademyofSciences,underaccessionnumbersCRA001584,CRA001584,thatarepubliclyaccessibleathttp://bigd.
big.
ac.
cn/gsa.
ORCIDTiejunLihttps://orcid.
org/0000-0001-5983-2985REFERENCESAlly,M.
S.
,Tang,J.
Y.
,Joseph,T.
,Thompson,B.
,Lindgren,J.
,Raphael,M.
A.
,…Epstein,E.
H.
(2014).
Theuseofvismodegibtoshrinkkeratocys‐ticodontogenictumorsinpatientswithbasalcellnevussyndrome.
JAMADermatology,150(5),542–545.
https://doi.
org/10.
1001/jamadermatol.
2013.
7444Bonilla,X.
,Parmentier,L.
,King,B.
,Bezrukov,F.
,Kaya,G.
,Zoete,V.
,…Nikolaev,S.
I.
(2016).
Genomicanalysisidentifiesnewdriversandprogressionpathwaysinskinbasalcellcarcinoma.
NatureGenetics,48(4),398–406.
https://doi.
org/10.
1038/ng.
3525Cibulskis,K.
,Lawrence,M.
S.
,Carter,S.
L.
,Sivachenko,A.
,Jaffe,D.
,Sougnez,C.
,…Getz,G.
(2013).
Sensitivedetectionofsomaticpointmutationsinimpureandheterogeneouscancersamples.
NatureBiotechnology,31(3),213–219.
https://doi.
org/10.
1038/nbt.
2514FIGURE3Distributionpatternof71PTCH1mutationsrelatedtothedifferentdomainsofthepatchedprotein.
ThethicklineindicatestheSSD[Colourfigurecanbeviewedatwileyonlinelibrary.
com]|1607Frana,J.
A.
,deSousa,S.
F.
,Diniz,M.
G.
,Pereira,T.
D.
S.
F.
,deResende,T.
A.
C.
,Santos,J.
N.
D.
,…Gomes,C.
C.
(2018).
AbsenceofBRAFV600Emutationinodontogenickeratocysts.
JournalofOralPathologyandMedicine,47(2),186–191.
https://doi.
org/10.
1111/jop.
12671Izzi,L.
,Lévesque,M.
,Morin,S.
,Laniel,D.
,Wilkes,B.
C.
,Mille,F.
,…Charron,F.
(2011).
BocandGas1eachformdistinctShhreceptorcomplexeswithPtch1andarerequiredforShh‐mediatedcellprolifer‐ation.
DevelopmentalCell,20(6),788–801.
https://doi.
org/10.
1016/j.
devcel.
2011.
04.
017Li,H.
,&Durbin,R.
(2009).
FastandaccurateshortreadalignmentwithBurrows‐Wheelertransform.
Bioinformatics,25(14),1754–1760.
https://doi.
org/10.
1093/bioinformatics/btp324Li,T.
J.
(2011).
Theodontogenickeratocyst:Acyst,oracysticneo‐plasmJournalofDentalResearch,90(2),133–142.
https://doi.
org/10.
1177/0022034510379016Lindstrom,E.
,Shimokawa,T.
,Toftgard,R.
,&Zaphiropoulos,P.
G.
(2006).
PTCHmutations:Distributionandanalyses.
HumanMutation,27(3),215–219.
https://doi.
org/10.
1002/humu.
20296Madras,J.
,&Lapointe,H.
(2008).
Keratocysticodontogenictumour:Reclassificationoftheodontogenickeratocystfromcysttotumour.
Journal/CanadianDentalAssociation,74(2),165–165h.
McKenna,A.
,Hanna,M.
,Banks,E.
,Sivachenko,A.
,Cibulskis,K.
,Kernytsky,A.
,…DePristo,M.
A.
(2010).
TheGenomeAnalysisToolkit:AMapReduceframeworkforanalyzingnext‐generationDNAsequencingdata.
GenomeResearch,20(9),1297–1303.
https://doi.
org/10.
1101/gr.
107524.
110Miller,N.
M.
,Campbell,C.
M.
,&Deas,D.
E.
(2011).
Reconstructivesurgi‐calmanagementofarecurrentodontogenickeratocyst:Casereportand1‐yearfollow‐up.
TheJournaloftheMichiganDentalAssociation,93(9),38–43.
Morgan,T.
A.
,Burton,C.
C.
,&Qian,F.
(2005).
Aretrospectivereviewoftreatmentoftheodontogenickeratocyst.
JournalofOralandMaxillofacialSurgery,63(5),635–639.
https://doi.
org/10.
1016/j.
joms.
2004.
07.
026Partridge,M.
,&Towers,J.
F.
(1987).
Theprimordialcyst(odontogenickeratocyst):Itstumour‐likecharacteristicsandbehaviour.
TheBritishJournalofOral&MaxillofacialSurgery,25(4),271–279.
Qu,J.
,Yu,F.
,Hong,Y.
,Guo,Y.
,Sun,L.
,Li,X.
,…Li,T.
(2015).
UnderestimatedPTCH1mutationrateinsporadickeratocysticodontogenictumors.
OralOncology,51(1),40–45.
https://doi.
org/10.
1016/j.
oraloncology.
2014.
09.
016Rudin,C.
M.
(2012).
Vismodegib.
ClinicalCancerResearch:AnOfficialJournaloftheAmericanAssociationforCancerResearch,18(12),3218–3222.
https://doi.
org/10.
1158/1078-0432.
CCR-12-0568Stone,D.
M.
,Hynes,M.
,Armanini,M.
,Swanson,T.
A.
,Gu,Q.
,Johnson,R.
L.
,…Rosenthal,A.
(1996).
Thetumour‐suppressorgenepatcheden‐codesacandidatereceptorforSonichedgehog.
Nature,384(6605),129–134.
https://doi.
org/10.
1038/384129a0Stransky,N.
,Egloff,A.
M.
,Tward,A.
D.
,Kostic,A.
D.
,Cibulskis,K.
,Sivachenko,A.
,…Grandis,J.
R.
(2011).
Themutationallandscapeofheadandnecksquamouscellcarcinoma.
Science,333(6046),1157–1160.
https://doi.
org/10.
1126/science.
1208130Tenzen,T.
,Allen,B.
L.
,Cole,F.
,Kang,J.
S.
,Krauss,R.
S.
,&McMahon,A.
P.
(2006).
ThecellsurfacemembraneproteinsCdoandBocarecomponentsandtargetsoftheHedgehogsignalingpathwayandfeedbacknetworkinmice.
DevelopmentalCell,10(5),647–656.
https://doi.
org/10.
1016/j.
devcel.
2006.
04.
004Toftgard,R.
(2000).
Hedgehogsignallingincancer.
CellularandMolecularLifeSciences,57(12),1720–1731.
https://doi.
org/10.
1007/PL00000654Wang,Y.
,Song,F.
,Zhu,J.
,Zhang,S.
,Yang,Y.
,Chen,T.
,…Zhao,W.
(2017).
GSA:Genomesequencearchive*.
GenomicsProteomicsBioinformatics,15(1),14–18.
https://doi.
org/10.
1016/j.
gpb.
2017.
01.
001Yang,H.
,Robinson,P.
N.
,&Wang,K.
(2015).
Phenolyzer:Phenotype‐basedprioritizationofcandidategenesforhumandiseases.
NatureMethods,12(9),841–843.
https://doi.
org/10.
1038/NMETH.
3484Yao,S.
,Lum,L.
,&Beachy,P.
(2006).
Theihogcell‐surfaceproteinsbindHedgehogandmediatepathwayactivation.
Cell,125(2),343–357.
https://doi.
org/10.
1016/j.
cell.
2006.
02.
040Yap,K.
L.
,Kiyotani,K.
,Tamura,K.
,Antic,T.
,Jang,M.
,Montoya,M.
,…Nakamura,Y.
(2014).
Whole‐exomesequencingofmuscle‐invasivebladdercanceridentifiesrecurrentmutationsofUNC5Candprog‐nosticimportanceofDNArepairgenemutationsonsurvival.
ClinicalCancerResearch:AnOfficialJournaloftheAmericanAssociationforCancerResearch,20(24),6605–6617.
https://doi.
org/10.
1158/1078-0432.
CCR-14-0257Yu,F.
Y.
,Hong,Y.
Y.
,Qu,J.
F.
,Chen,F.
,&Li,T.
J.
(2014).
Thelargein‐tracellularloopofptch1mediatesthenon‐canonicalHedgehogpathwaythroughcyclinB1innevoidbasalcellcarcinomasyndrome.
InternationalJournalofMolecularMedicine,34(2),507–512.
https://doi.
org/10.
3892/ijmm.
2014.
1783BIGDataCenterMembers(2019).
DatabaseresourcesoftheBIGDataCenterin2019.
NucleicAcidsResearch,47(D1),D8–D14.
https://doi.
org/10.
1093/nar/gky993SUPPORTINGINFORMATIONAdditionalsupportinginformationmaybefoundonlineintheSupportingInformationsectionattheendofthearticle.
Howtocitethisarticle:QuJ,ZhangJ,ZhangH,etal.
PTCH1alterationsarefrequentbutothergeneticalterationsarerareinsporadicodontogenickeratocysts.
OralDis.
2019;25:1600–1607.
https://doi.
org/10.
1111/odi.
13135
腾讯云轻量服务器两款低价年付套餐 2核4GB内存8M带宽 年74元
昨天,有在"阿里云秋季促销活动 轻量云服务器2G5M配置新购年60元"文章中记录到阿里云轻量服务器2GB内存、5M带宽一年60元的活动,当然这个也是国内机房的。我们很多人都清楚备案是需要接入的,如果我们在其他服务商的域名备案的,那是不能解析的。除非我们不是用来建站,而是用来云端的,是可以用的。这不看到其对手腾讯云也有推出两款轻量服务器活动。其中一款是4GB内存、8M带宽,这个比阿里云还要狠。这个真...
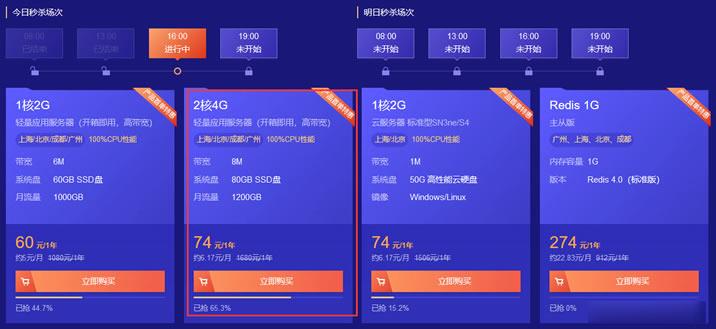
SugarHosts糖果主机圣诞节促销 美国/香港虚拟主机低至6折
SugarHosts 糖果主机商我们算是比较熟悉的,早年学会建站的时候开始就用的糖果虚拟主机,目前他们家还算是为数不多提供虚拟主机的商家,有提供香港、美国、德国等虚拟主机机房。香港机房CN2速度比较快,美国机房有提供优化线路和普通线路适合外贸业务。德国欧洲机房适合欧洲业务的虚拟主机。糖果主机商一般是不会发布黑五活动的,他们在圣圣诞节促销活动是有的,我们看到糖果主机商发布的圣诞节促销虚拟主机低至6折...
Gcorelabs:美国GPU服务器,8路RTX2080Ti;2*Silver-4214/256G内存/1T SSD,1815欧/月
gcorelabs怎么样?gcorelabs是创建于2011年的俄罗斯一家IDC服务商,Gcorelabs提供优质的托管服务和VPS主机服务,Gcorelabs有一支强大的技术队伍,对主机的性能和稳定性要求非常高。Gcorelabs在 2017年收购了SkyparkCDN并提供全球CDN服务,目标是进入全球前五的网络服务商。G-Core Labs总部位于卢森堡,在莫斯科,明斯克和彼尔姆设有办事处。...
meizhai为你推荐
-
支持ipad奶粉ios8支持ipad孩子apple平台操作使用手册css3圆角在HTML里如何实现圆角矩形?netbios端口如何组织netbios端口的外部通信勒索病毒win7补丁win7有针对勒索病毒的补丁吗itunes备份itunes 里面的资料如何备份?win7telnet怎样开启Windows7系统中的Telnet服务