Smythwww.07ppp.com
www.07ppp.com 时间:2021-04-08 阅读:()
RESEARCHOpenAccessAfilter-basedfeatureselectionapproachforidentifyingpotentialbiomarkersforlungcancerIn-HeeLee,GeraldHLushington*andMaheshVisvanathan*AbstractBackground:Lungcanceristheleadingcauseofdeathfromcancerintheworldanditstreatmentisdependantonthetypeandstageofcancerdetectedinthepatient.
Molecularbiomarkersthatcancharacterizethecancerphenotypearethusakeytoolinplanningatherapeuticresponse.
Acommonprotocolforidentifyingsuchbiomarkersistoemploygenomicmicroarrayanalysistofindgenesthatshowdifferentialexpressionaccordingtodiseasestateortype.
Data-miningtechniquessuchasfeatureselectionareoftenusedtoisolate,fromamongalargemanifoldofgeneswithdifferentialexpression,thosespecificgeneswhosedifferentialexpressionpatternsareofoptimalvalueinphenotypicdifferentiation.
Onesuchtechnique,BiomarkerIdentifier(BMI),hasbeendevelopedtoidentifyfeatureswiththeabilitytodistinguishbetweentwodatagroupsofinterest,whichisthushighlyapplicableforsuchstudies.
Results:MicroarraydatawithvalidatedgeneswasusedtoevaluatetheutilityofBMIinidentifyingmarkersforlungcancer.
Thisdatasetcontainsasetof129geneexpressionprofilesfromlarge-airwayepithelialcells(60samplesfromsmokerswithlungcancerand69fromsmokerswithoutlungcancer)and7genesfromthisdatahavebeenconfirmedtobedifferentiallyexpressedbyquantitativePCR.
Usingthisdataset,BMIwascomparedwithvariouswell-knownfeatureselectionmethodsandwasfoundtobemoresuccessfulthanothermethodsinfindingusefulgenestoclassifycanceroussamples.
AlsoitisevidentthatgenesselectedbyBMI(giventhesamenumberofgenesandclassificationalgorithms)showedbetterdiscriminativepowerthanthosefromtheoriginalstudy.
AfterpathwayanalysisontheselectedgenesbyBMI,wehavebeenabletocorrelatetheselectedgeneswithwell-knowncancer-relatedpathways.
Conclusions:OurresultsshowthatBMIcanbeusedtoanalyzemicroarraydataandtofindusefulgenesforclassifyingsamples.
PathwayanalysissuggeststhatBMIissuccessfulinidentifyingbiomarker-qualitycancer-relatedgenesfromthedata.
BackgroundLungcanceraccountsforlargeportionofcancerdeaths(29%)intheUnitedStatesformenaswellaswoman[1].
Themajortypesoflungcanceraresmall-cellandnon-small-cellcancer.
Non-small-cellcancercanbefurtherdividedintothreehistologicalsubtypes:squa-mous-cellcarcinoma,adenocarcinomaandlargecelllungcancer[2].
Regardlessofsubtype,the5-yearsurvi-valrateforlungcancerisamongthelowestofallcan-cersat15%(dataforUSA)[1].
Sincethetreatmentoflungcancerdependsonthesubtypeandthestageofcancer,itisimportanttohavedeterminedspecificmole-cularbiomarkersthatcanidentifythetypeofcancerasafunctionofgenescloselyrelatedtoeachdistinctphenotype.
Withadvanceofmicroarraytechnologies,itispossibletoconducthighthroughputdeterminationoftherela-tiverateswithwhichgenesareexpressedinagivencellortissuetype.
Thiscanhelpresearchersbetterunder-standadiseaseatthegenomiclevelandhasbecomeanimportanttoolinbiologicalsciencesaswellasmedicalandpharmaceuticalresearch.
Inthecontextoflungcan-cer,microarraytechnologycanbeusedtoidentifygeneswhoseexpressionprofileinatypeofcancerdiffersfromnormaltissuesorfromothertypesofcancer.
Suchbiomarkersareimportantsincetheycanprovidethebasisforimprovingadiagnosticclassifierorforenhan-cingthepredictionofpatient-specificprognosisor*Correspondence:glushington@ku.
edu;mvisvanathan@ku.
eduBioinformaticsCoreFacility,UniversityofKansas,Lawrence,KS66046,USALeeetal.
JournalofClinicalBioinformatics2011,1:11http://www.
jclinbioinformatics.
com/content/1/1/11JOURNALOFCLINICALBIOINFORMATICS2011Leeetal;licenseeBioMedCentralLtd.
ThisisanOpenAccessarticledistributedunderthetermsoftheCreativeCommonsAttributionLicense(http://creativecommons.
org/licenses/by/2.
0),whichpermitsunrestricteduse,distribution,andreproductioninanymedium,providedtheoriginalworkisproperlycited.
therapeuticresponse[3].
Fromaninformaticsperspec-tive,theprocessofselectingdifferentiallyexpressedgenesisreadilyachievedviadata-miningtechniquesknownasfeatureselection.
Featureselection,animpor-tantstepinthedata-miningprocess,aimstofindrepre-sentativefeaturesubsetsthatmeetdesiredcriteria.
Inmicroarraydataanalysis,onecriterionforadesiredfea-turesubsetwouldbeasetofgeneswhoseexpressionpatternsvarysignificantlywhencomparedacrossdiffer-entsamplegroups.
Theresultingsubsetcanthenbeusedtofurtheranalysissuchasbuildingadiagnosticclassifier.
Featureselectionmethods,ingeneral,canbecategor-izedintothreetypes,dependingonhowtheyarecom-binedwithotheranalysissteps:filtermethods,wrappermethodsandembeddedmethods[4].
Filtermethodsassesstherelevanceoffeaturesasscoresbylookingonlyatthepropertiesofthedata.
Featurescanbesortedbytheirscoresandlow-scoringfeaturescanberemoved.
Wrappermethodsembedtheanalysismodelwithinthefeaturesubsetsearch.
Inthissetup,asubsetoffeaturesisevaluatedbyapplyingaspecificanalysismodeltoreduceddatawiththeselectedfeaturesubset.
Inembeddedmethods,thesearchforanoptimalfeaturesubsetisbuiltintotheanalysisalgorithm.
Filtermethodsarethemostcommonlyappliedinbioinformaticsstu-diessincetheyarecomputationallysimple,fastandindependentofotheranalysisalgorithms.
Alsotheyallowfeaturestobequantifiedandprioritizedaccordingtothescores,whichisparticularlyimportantforbiologi-calinterpretation.
Inthispaper,afilter-basedfeatureselectionmethod,biomarkeridentifier(BMI),isadoptedtoanalyzegeneexpressiondatathatmightbeusedtodiscriminatebetweensampleswithandwithoutlungcancer.
Thedataconsistsofgeneexpressionpatternsinhistologi-callynormallarge-airwayepithelialcellsobtainedviabronchoscopyfromsmokers.
Genesidentifiedusingthisdatasetcanbeusedtodiagnosinglungcanceramongsmokerswithsuspectedlungcancer.
ThegenesselectedbyBMIwerecomparedwiththosefromvariousotherfeatureselectionalgorithmsandthoseidentifiedfromtheoriginalexperimentalstudy.
PathwayanalysisforthegenesselectedbyBMIwasalsoperformed.
MethodsBiomarkerIdentifierThebiomarkeridentifier(BMI)[5,6]methodcombinesvariousstatisticalmeasurestodiscerntheabilityoffea-turestodistinguishbetweentwodatagroupsofinterest.
Itconsidersthreemeasuresforevaluatingfeatures.
First,itcheckswhetherdistributionofafeatureissignificantlydifferentbetweendatagroups.
Ifthedistributionofafeaturechangessubstantially,thefeaturemightberelevanttotheunderlyingdifferencebetweendatagroups.
Second,theratioofoverallvariancerelativetovarianceincontrolgroupisusedtomeasuretherelia-bilityofafeature.
Forexample,iftheoverallvarianceisgreaterthanthatofcontrolgroup,itmeansthatthefea-turedisplaysmorenoisybehaviorinexperimentgroupmakingitlessusefulunlessitalsodemonstratesasignif-icantchangebetweendatagroup.
Ontheotherhand,anoverallvariancesmallerthanthatofcontrolgroupimpliesthatthefeatureshowsmoreconsistentbehaviorintheexperimentgroup,makingitamoreusefulfea-tureprovidedthatthereexistsasignificantdifferencebetweenthecontrasteddatagroups.
Forthesereasons,BMIpenalizesorcreditsascoreofafeaturebytheratioofoverallvariancerelativetovarianceincontrolgroup.
Lastly,BMIconsidersthediscriminativepowerofeachindividualfeaturebyincorporatingthetruepositiveratefromlogisticregressionusingthefeature.
Inmathemati-calterms,letusassumeadatasetDconsistingoftwogroups'control(ctr)'and'experiment(exp)'.
BMIassignsascoreforafeaturexdefinedasfollows:BMI(x)=λ·TP2|diff|CVctrCV,wherediff=,if≥11,otherwise.
Here,lisascalingfactorandTP2istheproductofthetruepositive(TP)ratesdeterminedforeachgroupsusinglogisticregressionoftheform'outcome~feature'.
CVctrandCVdenotethecoefficientofvarianceforthefeaturexinthe'control'groupandinbothgroups,respectively.
Also,Δ=x/xctr,wherexctr,andxdenotethemeanvalueofxin'control'andinbothgroups,respectively.
Forbiologicaldatasuchasmicroarray,thesignofΔdiffforaparticulargenecanbeinterpretedasover-expressionorunder-expressionin'experiment'comparedto'control';positiveasover-expressionandnegativeasunder-expression.
BMIhasshownpromisingresultsonvariousdatasetssuchasmassspectrometrydataofmetabolites[5],liverdisease[7]andmicroarraydatafromvarioustypesofcancer[6].
Inthisstudy,itisusedtoidentifypotentialbiomarkersforlungcancerfrommicroarraydata.
OtherfeatureselectionmethodsForcomparisonwithBMI,weused6differentpopularfeatureselectionmethods:informationgain(IG),Relief-F(RF),t-test(T)anditstwovariants(moder-atedt-test(MT)andwindowt-test(WT)),andchi-squaredtest(CS).
Leeetal.
JournalofClinicalBioinformatics2011,1:11http://www.
jclinbioinformatics.
com/content/1/1/11Page2of8InformationgainInformationgain(relativeentropy,orKullback-Leiblerdivergence),inprobabilitytheoryandinformationthe-ory,isameasureofthedifferencebetweentwoprob-abilitydistributions.
Itevaluatesafeaturexbymeasuringtheamountofinformationgainedwithrespecttotheclass(orgroup)variabley,definedasfol-lows:I(x)=H(P(y))H(P(y|x)).
Specifically,itmeasuresthedifferencebetweenthemarginaldistributionofobservableyassumingthatitisindependentoffeaturex(P(y))andtheconditionaldis-tributionofyassumingthatitisdependentofx(P(y|x)).
Ifxisnotdifferentiallyexpressed,ywillbeindepen-dentofx,thusxwillhavesmallinformationgainvalue,andviceversa.
Relief-FRelief-F[8]isaninstance-basedfeatureselectionmethodwhichevaluatesafeaturebyhowwellitsvaluedistinguishessamplesthatarefromdifferentgroupsbutaresimilartoeachother.
Foreachfeaturex,Relief-Fselectsarandomsampleandkofitsnearestneighborsfromthesameclassandeachofdifferentclasses.
Thenxisscoredasthesumofweighteddifferencesindiffer-entclassesandthesameclass.
Ifxisdifferentiallyexpressed,itwillshowgreaterdifferencesforsamplesfromdifferentclasses,thusitwillreceivehigherscore(orviceversa).
t-testandvariantsTheStudent'st-test[9]istraditionallyusedtocomparetwonormallydistributedsamplesorpopulations.
Itpre-fersfeatureswithamaximaldifferenceofmeanvaluebetweengroupsandaminimalvariabilitywithineachgroup,butitcanfailwhentherearesmallnumberofsamplesortheestimatedvariancesarenotequalbetweengroups(heteroscedasticity):scenarioswhicharecommonforpracticaldata.
Tocopewithsuchpro-blems,Welchproposedavariantoft-testtakinghetero-scedasticityintoaccount[10].
VariousstatisticaltestsfordifferentialexpressionarebasedonthetraditionalStudentandWelchtests.
Smyth[11]appliedahierarch-icalBayesianapproach(moderatedt-test)totheStudentandWelchtestsandintegratedmoreaprioriinforma-tiontoyieldmorerobustestimates.
Bergeretal.
[12]suggestedawindowt-testthatusesmultiplegeneswhichshareasimilarexpressionleveltocomputethevariancetobeincorporatedinthet-test.
Inthiswork,wechoseWelch'st-test,moderatedt-testandwindowt-testforcomparison.
chi-squaredtestChi-squaredtestisanotherpopularstatisticaltestofthedivergencebetweentheobservedandexpecteddistribu-tionofafeature.
Infeatureselection,ittestswhetherthedistributionofafeaturediffersbetweengroups.
Thechi-squarescoreusesthesummationofsquareddiffer-encesbetweenobservedandexpectedvaluesdividedbyexpectedvalues.
ExperimentaldataSpiraetal.
reportedgeneexpressiondatafromlargeair-wayepithelialcellsbymicroarrayanalysis[13].
Thisdatasetcoversasetof129AffymetrixHG-U133Amicroar-rayscomparing60smokerswithlungcancerand69smokerswithoutlungcancer.
Thisexperimentwasdesignedtodetermineifgeneexpressioninhistologi-callynormallarge-airwayepithelialcellsobtainedviabronchoscopyfromsmokerswithsuspectedlungcancercouldbeusedasalungcancerbiomarker.
Inthisdataset,7geneswereconfirmedtobedifferentiallyexpressedbetweencanceroussamplesandnon-cancer-oussamplesbyquantitativePCR[13].
TheRobustMul-tichipAverage(RMA)algorithm[14]wasusedforbackgroundadjustment,normalization,andprobe-levelsummarizationofthemicroarraysamples(pleaserefertosupplementarymethodsof[13]fordetailedinforma-tion).
Thedatasetcanbeaccessedfromgeneexpres-sionomnibus(GEO,http://www.
ncbi.
nlm.
nih.
gov/geo/)underaccessionnumberofGSE4115.
ThisdatasetwaschosensinceitconsistedofasignificantnumberofreplicatesandsomeofthegenesinthedatasetwereconfirmedbyquantitativePCR,whichprovidesagoodbasisforpreliminaryvalidation.
Tocontrastperformanceamongfeatureselectionmethods,wealsousedthedatasetpublishedthroughMicroArrayQualityControlprojectphaseII(MAQC-II).
Among9non-controldatasetsfromMAQC-II,thedatasetwiththemostbalancednumberofpositive/negativesamples(breastcancerdatawithestrogenreceptorstatusasclass)waschosen.
Thedatasetcon-sistsoftraining(130samples)andvalidation(100sam-ples)sets.
TheprocesseddatawasobtainedthroughGEOunderaccessionnumberGSE20194.
ResultsandDiscussionComparisonwithotherfeatureselectionmethodsFeatureselectionmethodscanbeevaluatedinvariousways.
Onepopularwayistoobservetheclassificationperformanceusingthefeaturesselectedbythemethod.
Ifafeatureselectionmethodisabletochoosetrulysig-nificantfeatures,theclassifiertrainedusingthosefea-turesshouldshowgoodperformancewithasmallnumberoffeatures.
Ifimportantfeaturesarealreadyknown,ontheotherhand,wecanevaluatefeatureselectionmethodsbyhowtheyrankthoseknownfea-tures.
SinceimportantfeatureshavenotbeenreportedfortheMAQC-IIdataset,itcanbeapproachedonlyviathefirstevaluationstrategy,buttheairwaydatasetisLeeetal.
JournalofClinicalBioinformatics2011,1:11http://www.
jclinbioinformatics.
com/content/1/1/11Page3of8amenabletobothmodesofevaluationsincesomeofgeneshavebeenexperimentallyconfirmedtobediffer-entiallyexpressed.
SinceaseparatevalidationsetisavailablewithintheMAQC-IIdata,weusedthetrainingsetforfeatureselec-tionandvalidationsetforclassification.
Thatis,featureselectionmethodsarefirstappliedtotrainingsettoobtainfeaturesubsets.
Then,foreachfeatureselectionmethod/classificationalgorithmpairing,classificationperformancesareevaluatedonthevalidationsetthrough10-foldcross-validationwithvaryingnumberoffeatures(from1to60).
AUCvalues(areaunderthecurve;apop-ularmeasureformodelcomparisoninmachinelearningresearchinterpretedastheprobabilitythat,givenaran-domlypickedpositiveexampleandnegativeexample,theclassifierwillassignahigherscoretothepositiveexam-plethantothenegativeone)havebeenusedhereintomeasureclassificationperformance.
LargerAUCvaluesimplymorepreciseclassification.
Forimplementation,weusedWeka[15],apopularmachinelearninglibrarywritteninJava,andthedefaultsettingwasusedforeachclassificationalgorithm.
Table1showsthemaximumAUCvalueachievedbyeachcombinationoffeatureselectionmethodsandclassificationalgorithmsfortheMAQC-IIdataset.
WecanseethattheclassifiersincombinationwithBMIshowperformancelevelscompar-abletootherswithrelativelysmallnumberoffeatures.
Also,thefeaturesselectedbyBMIshowstableperfor-manceregardlesstheclassificationalgorithm.
Fortheairwaydataset,weappliedasimilarten-foldcross-validationapproachaswiththeMAQC-IIdatatocompareclassificationperformanceofdifferentfeatureselectionmethods.
Here,thedatawasdividedinto10-folds,whereby9foldsareusedforbothselectingfeaturesandtrainingclassifiers,andthereservedfoldwasusedtocalculateAUCvalueoftrainedclassifiers.
Foreachcombi-nationoffeatureselectionmethodsandclassificationalgo-rithms,thisprocesswasrepeated10timeswithadifferentreservedfold,whilevaryingnumberoffeatures(from1to60)andtheAUCvalueswereaveragedoverthetendis-tinctreserved-foldcases.
TheparametersettingforeachclassificationalgorithmwasthesameasinMAQC-IIdataset.
Table2showsthemaximumAUCvalueachievedbyeachcombinationoffeatureselectionmethodsandclassi-ficationalgorithms.
AsinMAQC-IIdataset,theclassifiersincombinationwithBMIshowcomparableperformancewithotherswithrelativelysmallnumberoffeatures.
AndthefeaturesselectedbyBMIshowstableperformanceregardlesstheclassificationalgorithm.
Next,fortheairwaydataset,weinvestigatedhowthegenesconfirmedintheliterature(DUOX1,BACH2,DCLRE1C,RAB1A,TPD52,FOS,andIL8)arerankedbyBMIcomparedtootherfeatureselectionmethods.
Ifthesegenesaregenerallyrankedhighly,afeatureTable1ComparisonofclassificationperformancesonMAQC-IIdatasetClassificationAlgorithmsFeatureSelectionMethodsSupportVectorMachinek-NearestNeighborNaiveBayesRandomForestInformationGain0.
9031(6)0.
9380(25)0.
9008(40)0.
9206(50)Chi-squaredtest0.
8821(1)0.
9164(50)0.
9151(4)0.
9441(60)Relief-F0.
8821(1)0.
9052(15)0.
8995(50)0.
9306(60)t-test0.
9067(15)0.
9100(20)0.
9042(8)0.
9304(40)Windowt-test0.
8903(5)0.
9216(5)0.
9012(2)0.
9199(10)Moderatedt-test0.
8903(6)0.
9084(5)0.
8987(1)0.
9309(50)BMI0.
9077(4)0.
9298(15)0.
9164(4)0.
9250(9)EachvaluerepresentsthemaximumAUCvalue(by10-foldcross-validation)achievedbythecorrespondingfeatureselectionmethodandclassificationalgorithm.
Thenumberoffeaturesusedtoachievethemaximumisshowninsideparenthesis.
Table2ComparisonofclassificationperformancesonairwaydatasetClassificationAlgorithmsFeatureSelectionMethodsSupportVectorMachinek-NearestNeighborNaiveBayesRandomForestInformationGain0.
6853(40)0.
8006(4)0.
8297(50)0.
8620(60)Chi-squaredtest0.
7052(20)0.
8029(60)0.
7997(3)0.
8309(50)Relief-F0.
6633(25)0.
7825(9)0.
8329(25)0.
8685(60)t-test0.
6902(8)0.
7822(4)0.
8402(4)0.
8121(8)Windowt-test0.
6856(20)0.
7817(30)0.
8367(20)0.
8093(40)Moderatedt-test0.
6878(6)0.
7875(5)0.
8329(5)0.
8115(20)BMI0.
7572(9)0.
8005(5)0.
8299(5)0.
8212(10)EachvaluerepresentsthemaximumAUCvalue(via10-foldcross-validation)achievedbythecorrespondingfeatureselectionmethodandclassificationalgorithm.
Thenumberoffeaturesusedtoachievethemaximumisshowninsideparenthesis.
Leeetal.
JournalofClinicalBioinformatics2011,1:11http://www.
jclinbioinformatics.
com/content/1/1/11Page4of8selectionmethodcouldbesaidtocorroboratethegivendata.
Asbefore,wedividedthedatainto10foldsandusedonly9foldsinfeatureselection,repeatingthefea-tureselectionforeachdistinctreservedfold.
Foreachofthesetenfoldcases,werecordedgeneranksasdeter-minedbyeachmethodandcalculatedthemedianvalueforeachgene.
Figure1showsmedianranksofvalidatedgenesbydifferentfeatureselectionmethods,demonstrat-ingthatBMIranksalloftheconfirmedgeneswithinthetop4000rankedgenes,andtheoverallBMIrankingofconfirmedgenesisgenerallysuperiortoothermethods.
Fromtheseresults,itcanbesaidthatBMIshowscompetitiveperformanceinidentifyingusefulfeaturesforclassificationandshowshighconsistencywithactualdifferentialexpression.
ComparisonwithbiomarkersfromliteratureFortheairwaydataset,wefurthercomparedthegenesselectedbyBMIandthebiomarkersfromoriginalliterature[13].
Inoriginalliterature,80featureswereselectedtodistinguishcanceroussamplesfromnormalsamples.
ForBMI,wechose10featuresthatwereusedtoachievethebestclassificationperformanceinTable2.
Theselected10fea-turesareshowninTable3.
Thenwetrainedvariouspopularclassificationalgorithmsusingthesetwosetsoffeatures:naveBayes,supportvectormachine(SVM),neuralnetwork,k-nearestneighbor,andrandomforest.
Weusedtheimple-mentationinWekasoftware[15]withdefaultsettings.
Table4showsthedetailedclassificationperformancesobtainedfrom20independentrunsof10-foldcross-validation.
ClassifierstrainedusingfeaturesselectedbyBMIgenerallyshowedbetterperformanceformostclas-sificationalgorithms.
ThisimpliesthatthefeaturesselectedbyBMIaremoreusefulforconstructingaccu-rateclassifiers,whichcanprovideagoodbasisforfurtherscreeningofbiomarkers.
PathwayanalysisofselectedbiomarkersAlthoughasetofgenesisusefulfortrainingclassifier,theconstituentgenesmaybeuselessasbiomarkersifFigure1Themedianranksofvalidatedgenesinairwaydatasetbyvariousfeatureselectionmethods.
Table3Top10genesselectedbyBMIProbeIDSymbolRegulationName201694_s_atEGR1Upearlygrowthresponse1202056_atKPNA1Upkaryopherinalpha1(importinalpha5)203265_s_atMAP2K4Upmitogen-activatedproteinkinasekinase4207283_atRPL23AP13DownribosomalproteinL23apseudogene13211612_s_atIL13RA1Upinterleukin13receptor,alpha1214261_s_atADH6Upalcoholdehydrogenase6(classV)216609_atTXNDownFulllengthinsertcDNAcloneYI46D09219233_s_atGSDMBDowngasderminB222339_x_at-Down-34206atARAP1DownArfGAPwithRhoGAPdomain,ankyrinrepeatandPHdomain1Leeetal.
JournalofClinicalBioinformatics2011,1:11http://www.
jclinbioinformatics.
com/content/1/1/11Page5of8theirbiologicalrolesarenotrelatedtothetargetdiseaseorprocess.
Thusweanalyzedthepathwaysassociatedwith80highly-rankedgenestoinvestigatetheirbiologi-calroles.
Forpathwayanalysis,weinvestigatedasso-ciatedtermsinKEGGpathways[16],NCI-Naturepathwayinteractiondatabase[17],andPANTHER(pro-teinanalysisthroughevolutionaryrelationships)classifi-cationsystem[18]usingtheEGANprogram[19].
Tables5and6summarizethegenesandtheirasso-ciatedpathwayswithsignificantp-values(<0.
05).
Wecanobservethattherearesomegenes(EGR1,FOS,DUSP10,andMAP2K4)associatedwithmitogen-acti-vatedproteinkinase(MAPK)pathways,whichisawell-knowntargetintheoncologydrugdiscovery[20].
Also,threegenes(APC,MSH2,andATF3)showedsignificantassociationwithatermfromtheNCI-NaturePathwayInteractionDatabase,'Directp53effectors.
'Thisimpliesthatthosegenesarerelatedwithprotein'p53'whichisknownasatumorsuppressorprotein[21].
WenotethatincidenceofthegeneralKEGGannotation'path-waysincancer'showedagoodassociation(p-valueof0.
0019)withoursetof80genes.
Onealsofindsotherpathwaysrelatedwithknownoncogenessuchasc-Met[22]andepidermalgrowthfactorreceptor(EGFRorErbB-1)[23]withinourlist.
Fromthese,itcanbesaidthatgeneshighlyrankedbyBMIaregenerallyrelevanttocancerdevelopmentordiagnosis,thusBMIappearstobeusefulforidentifyingpotentialbiomarkersforlungcancer.
ConclusionsInthiswork,afilter-basedfeatureselectionmethod,biomarkeridentifier(BMI),hasbeenappliedtofindpotentialbiomarkersforlungcancerfrommicroarrayTable4ClassificationperformanceswithselectedbiomarkersbyBMIandoriginalliteratureBiomarkersbyBMIBiomarkersfromoriginalliteratureClassifierSpecificitySensitivityAccuracySpecificitySensitivityAccuracyNaveBayes0.
7938++0.
7006++0.
7489++0.
71170.
66440.
6872SVM0.
8134++0.
7056++0.
7615++0.
66220.
65930.
6607NeuralNetwork0.
7242++0.
64220.
68480.
69560.
7459++0.
7217++k-NearestNeighbor0.
8325++0.
61440.
7275++0.
63780.
6964++0.
6682RandomForest0.
7139++0.
7328++0.
7230++0.
68720.
66800.
6773++and+denotessuperiorperformanceasdeterminedatof1%and5%significancelevelsrespectively.
Table5KEGGpathwaysandPANTHERclassificationsassociatedwithtop80genesselectedbyBMIKEGGpathwaynamep-valueAssociatedgenesColorectalcancer1.
3809E-4FOS,MSH2,APCPathwaysincancer0.
0019FOS,MSH2,APC,TCEB2Metabolicpathways0.
0021ADH6,SAT1,EXT2,TGDS,BTD,PRPS1,AGPSBiotinmetabolism0.
0032BTDMAPKsignalingpathway0.
0094DUSP10,MAP2K4,FOSCytokine-cytokinereceptorinteraction0.
0098CXCR4,ACVR2A,IL13RA1Toll-likereceptorsignalingpathway0.
0117FOS,MAP2K4Tightjunction0.
0196PPP2R2D,INADLMismatchrepair0.
0361MSH2Glycosaminoglycanbiosynthesis-heparansulfate0.
0408EXT2Pentosephosphatepathway0.
0423PRPS1Endocytosis0.
0428ARAP1,CXCR4PANTHERclassificationp-valueAssociatedgenesOxidativestressresponse8.
6417E-5TXN,MAP2K4,DUSP10O-antigenbiosynthesis0.
0064TGDSTcellactivation0.
0083FOS,B2MInterleukinsignalingpathway0.
0108IL13RA1,FOSApoptosissignalingpathway0.
0133ATF3,FOSFGFsignalingpathway0.
0135MAP2K4,PPP2R2DAxonguidancemediatedbySlit/Robo0.
0253CXCR4HypoxiaresponseviaHIFactivation0.
0408TXNInsulin/IGFpathway-mitogenactivatedproteinkinasekinase/MAPkinasecascade0.
0484FOSLeeetal.
JournalofClinicalBioinformatics2011,1:11http://www.
jclinbioinformatics.
com/content/1/1/11Page6of8data.
BMImeasuresthepotentialvalueofeachgeneasabiomarkercandidatebycombiningvariousstatisticalmeasurestoassessitsabilitytodistinguishbetweentwodatagroupsofinterest.
WeevaluatedBMIperformanceontwopublicmicroarraydatasets:onefromtheMicro-ArrayQualityControlprojectandtheotherfromsmo-kerswithandwithoutlungcancer.
BMIwascomparedwithotherpopularfilter-basedfeatureselectionmethodsonbothdatasetandshowedcompetitiveperformanceinselectingusefulfeaturesforvariousclassificationalgo-rithms.
SinceofthelatterdatasetincludesinformationregardingspecificgeneswhosetissuedifferentiationrelevancehasbeenvalidatedbyquantitativeRT-PCR,wealsocomparedhowthesegeneswererankedbydif-ferentfeatureselectionalgorithm.
ThevalidatedgenesgenerallywereassignedhigherranksbyBMIthanbyothermethods,implyingthatBMIshouldbeeffectiveinidentifyingbiomarkersthatshowdifferentialexpressionincanceroussamples.
WealsocomparedBMIwiththeapproachintheoriginalanalysisconductedonthelungcancermicroarraydata[13]bycontrastingtheclassifica-tionperformanceusingselectedgenesfromeachTable6NCI-Naturepathwayinteractionsassociatedwithtop80genesselectedbyBMINCI-NaturePathwayInteractionp-valueAssociatedgenesATF-2transcriptionfactornetwork6.
8276E-5ATF3,FOS,DUSP10DownstreamsignalinginnaveCD8+Tcells1.
8173E-4B2M,EGR1,FOSSignalingeventsmediatedbyHepatocyteGrowthFactorReceptor(c-Met)2.
6255E-4EGR1,MAP2K4,APCEphrinBreversesignaling8.
6116E-4CXCR4,MAP2K4ErbB1downstreamsignaling8.
7013E-4MAP2K4,FOS,EGR1Regulationofp38-alphaandp38-beta0.
0011DUSP10,MAP2K4Directp53effectors0.
0013APC,MSH2,ATF3TrkreceptorsignalingmediatedbytheMAPKpathway0.
0014EGR1,FOSRhoAsignalingpathway0.
0021FOS,MAP2K4IL6-mediatedsignalingevents0.
0023MAP2K4,FOSPresenilinactioninNotchandWntsignaling0.
0024FOS,APCCalcineurin-regulatedNFAT-dependenttranscriptioninlymphocytes0.
0025EGR1,FOSRegulationofAndrogenreceptoractivity0.
0027EGR1,MAP2K4Fc-epsilonreceptorIsignalinginmastcells0.
0041FOS,MAP2K4IL12-mediatedsignalingevents0.
0045B2M,FOSHIF-1-alphatranscriptionfactornetwork0.
0052FOS,CXCR4CDC42signalingevents0.
0058APC,MAP2K4RegulationofnuclearSMAD2/3signaling0.
0075FOS,ATF3Glucocorticoidreceptorregulatorynetwork0.
0077FOS,EGR1SumoylationbyRanBP2regulatestranscriptionalrepression0.
0174RANBP2JNKsignalingintheCD4+TCRpathway0.
0206MAP2K4RassignalingintheCD4+TCRpathway0.
0222FOSHypoxicandoxygenhomeostasisregulationofHIF-1-alpha0.
0284TCEB2CellularrolesofAnthraxtoxin0.
0346MAP2K4VEGFR3signalinginlymphaticendothelium0.
0361MAP2K4S1P2pathway0.
0377FOSPDGFR-alphasignalingpathway0.
0377FOSALK1signalingevents0.
0392ACVR2ASignalingeventsmediatedbyPRL0.
0392EGR1TRAILsignalingpathway0.
0438MAP2K4RegulationofCDC42activity0.
0453APCS1P3pathway0.
0453CXCR4CD40/CD40Lsignaling0.
0469MAP2K4CanonicalWntsignalingpathway0.
0469APCp38MAPKsignalingpathway0.
0469TXNCalciumsignalingintheCD4+TCRpathway0.
0484FOSNongenotropicAndrogensignaling0.
0484FOSNephrin/Neph1signalinginthekidneypodocyte0.
0499MAP2K4IL12signalingmediatedbySTAT40.
0499FOSLeeetal.
JournalofClinicalBioinformatics2011,1:11http://www.
jclinbioinformatics.
com/content/1/1/11Page7of8method.
Givenmodelstrainedforvariousclassificationalgorithms,classifiersbasedongenesselectedbyBMIshowedbetterperformancethanthosefromoriginalstudy.
Finally,inevaluatingwhetherthegenesselectedbyBMIhaveknownbiologicalfunctionrelatedto(lung)cancer,weanalyzedtheirpathwaydispositionandfoundthatmanygeneswereassociatedwithknowncancer-relatedpathways.
ThuswecanconcludethatBMIisasuitabletechniqueforphenotypicclassificationofmicro-arraydataandmayprovideareasonablemechanismforidentifyingviablediagnosticbiomarkercandidates.
Basedontheresultsinthisstudy,wearepursuingafol-low-upstudyusingBMItoidentifybiomarkerssuitableforthelungcanceranalysiswithexperimentaldataonclinicallyderivedtissues.
AcknowledgementsThispublicationwasmadepossiblebygrantnumberP20RR016475fromtheNationalCenterforResearchResources(NCRR),acomponentoftheNationalInstitutesofHealth(NIH).
WealsowouldliketothankDrs.
MichaelNetzerandChristianBaumgartnerfromUniversityofHealthSciences,MedicalInformaticsandTechnology(UMIT),AustriainprovidingsourcecodeforBMIimplementation.
Authors'contributionsILparticipatedinthedesignofthestudy,performedthestatisticalanalysisanddraftedthemanuscript.
GLandMVconceivedofthestudy,andparticipatedinitsdesignandcoordination.
Allauthorsreadandapprovedthefinalmanuscript.
CompetinginterestsTheauthorsdeclarethattheyhavenocompetinginterests.
Received:8October2010Accepted:21March2011Published:21March2011References1.
JemalA,SiegelR,WardE,HaoY,XuJ,MurrayT,ThunMJ:Cancerstatistics.
CACancerJClin2008,58:71-96.
2.
HerbstRS,HeymachJV,LippmanSM:Lungcancer.
NewEnglandJournalofMedicine2008,359:1367-1380.
3.
GranvilleCA,DennisPA:Anoverviewoflungcancergenomicsandproteomics.
AmericanJournalofRespiratoryCellandMolecularBiology2005,32:169-176.
4.
SaeysY,InzaI,LarraagaP:Areviewoffeatureselectiontechniquesinbioinformatics.
Bioinformatics2007,23:2507-2517.
5.
BaumgartnerC,BaumgartnerD:Biomarkerdiscovery,diseaseclassification,andsimilarityqueryprocessingonhigh-throughputMS/MSdataofinbornerrorsofmetabolism.
JournalofBiomolecularScreening2006,11:90-99.
6.
VisvanathanM,NetzerM,SegerM,AdagarlaBS,BaumgartnerC,SittampalamS,LushingtonGH:Oncogenesandpathwayidentificationusingfilter-basedapproachesbetweenvariouscarcinomatypesinlung.
InternationalJournalofComputationalBiologyandDrugDesign2009,2:236-251.
7.
NetzerM,MillonigG,OslM,PfeiferB,PraunS,VillingerJ,VogelW,BaumgartnerC:Anewensemble-basedalgorithmforidentifyingbreathgasmarkercandidatesinliverdiseaseusingionmoleculereactionmassspectrometry.
Bioinformatics2009,25(7):941-947.
8.
KononenkoI:Estimatingattributes:analysisandextensionsofRELIEF.
InECML-94:ProceedingsoftheEuropeanconferenceonmachinelearningonMachineLearning.
Editedby:BergadanoF,DeRaedtL.
SpringerBerlin/Heidelberg;1994:171-182.
9.
Student:Theprobableerrorofamean.
Biometrika1908,6:1-25.
10.
WelchBL:Thesignificanceofthedifferencebetweentwomeanswhenthepopulationvariancesareunequal.
Biometrika1938,29:350-362.
11.
SmythGK:LinearmodelsandempiricalBayesmethodsforassessingdifferentialexpressioninmicroarrayexperiments.
StatisticalApplicationsinGeneticsandMolecularBiology2004,3:3.
12.
BergerF,DeHertoghB,PierreM,GaigneauxA,DepiereuxE:The"Windowt-test":asimpleandpowerfulapproachtodetectdifferentiallyexpressedgenesinmicroarraydatasets.
CentralEuropeanJournalofBiology2008,3:327-344.
13.
SpiraA,BeaneJE,ShahV,SteilingK,LiuG,SchembriF,GilmanS,DumasYM,CalnerP,SebastianiP,SridharS,BeamisJ,LambC,AndersonT,GerryN,KeaneJ,LenburgME,BrodyJS:Airwayepithelialgeneexpressioninthediagnosticevaluationofsmokerswithsuspectlungcancer.
NatureMedicine2007,13:361-366.
14.
IrizarryRA,BolstadBM,CollinF,CopeLM,HobbsB,SpeedTP:SummariesofAffymetrixGeneChipprobeleveldata.
NucleicAcidsResearch2003,31:e15.
15.
HallM,FrankE,HolmesG,PfahringerB,ReutemannP,WittenIH:TheWEKADataMiningSoftware:AnUpdate.
Explorations2009,11:10-18.
16.
KanehisaM,GotoS,FurumichiM,TanabeM,HirakawaM:KEGGforrepresentationandanalysisofmolecularnetworksinvolvingdiseasesanddrugs.
NucleicAcidsResearch2010,38:D355-D360.
17.
SchaeferCF,AnthonyK,KrupaS,BuchoJ,DayM,HannayT,BuetowKH:PID:thepathwayinteractiondatabase.
NucleicAcidsResearch2009,37:D674-D679.
18.
ThomasPD,CampbellMJ,KejariwalA,MiH,KarlakB,DavermanR,DiemerK,MuruganujanA,NarechaniaA:PANTHER:alibraryofproteinfamiliesandsubfamiliesindexedbyfunction.
GenomeResearch2003,13:2129-2141.
19.
PaquetteJ,TokuyasuT:EGAN:exploratorygeneassociationnetworks.
Bioinformatics2010,26:285-286.
20.
Sebolt-LeopoldJS:AdvancesinthedevelopmentofcancertherapeuticsdirectedagainsttheRAS-mitogen-activatedproteinkinasepathway.
ClinicalCancerResearch2008,14:3651-3656.
21.
HollsteinM,SidranskyD,VogelsteinB,HarrisCC:p53mutationsinhumancancers.
Science1991,253:49-53.
22.
SattlerM,SalgiaR:c-Metandhepatocytegrowthfactor:Potentialasnoveltargetsincancertherapy.
CurrentOncologyReports2007,9:102-108.
23.
ZhangH,BerezovA,WangQ,ZhangG,DrebinJ,MuraliR,GreeneMI:ErbBreceptors:fromoncogenestotargetedcancertherapies.
TheJournalofClinicalInvestigation2007,117:2051-2058.
doi:10.
1186/2043-9113-1-11Citethisarticleas:Leeetal.
:Afilter-basedfeatureselectionapproachforidentifyingpotentialbiomarkersforlungcancer.
JournalofClinicalBioinformatics20111:11.
SubmityournextmanuscripttoBioMedCentralandtakefulladvantageof:ConvenientonlinesubmissionThoroughpeerreviewNospaceconstraintsorcolorgurechargesImmediatepublicationonacceptanceInclusioninPubMed,CAS,ScopusandGoogleScholarResearchwhichisfreelyavailableforredistributionSubmityourmanuscriptatwww.
biomedcentral.
com/submitLeeetal.
JournalofClinicalBioinformatics2011,1:11http://www.
jclinbioinformatics.
com/content/1/1/11Page8of8
Molecularbiomarkersthatcancharacterizethecancerphenotypearethusakeytoolinplanningatherapeuticresponse.
Acommonprotocolforidentifyingsuchbiomarkersistoemploygenomicmicroarrayanalysistofindgenesthatshowdifferentialexpressionaccordingtodiseasestateortype.
Data-miningtechniquessuchasfeatureselectionareoftenusedtoisolate,fromamongalargemanifoldofgeneswithdifferentialexpression,thosespecificgeneswhosedifferentialexpressionpatternsareofoptimalvalueinphenotypicdifferentiation.
Onesuchtechnique,BiomarkerIdentifier(BMI),hasbeendevelopedtoidentifyfeatureswiththeabilitytodistinguishbetweentwodatagroupsofinterest,whichisthushighlyapplicableforsuchstudies.
Results:MicroarraydatawithvalidatedgeneswasusedtoevaluatetheutilityofBMIinidentifyingmarkersforlungcancer.
Thisdatasetcontainsasetof129geneexpressionprofilesfromlarge-airwayepithelialcells(60samplesfromsmokerswithlungcancerand69fromsmokerswithoutlungcancer)and7genesfromthisdatahavebeenconfirmedtobedifferentiallyexpressedbyquantitativePCR.
Usingthisdataset,BMIwascomparedwithvariouswell-knownfeatureselectionmethodsandwasfoundtobemoresuccessfulthanothermethodsinfindingusefulgenestoclassifycanceroussamples.
AlsoitisevidentthatgenesselectedbyBMI(giventhesamenumberofgenesandclassificationalgorithms)showedbetterdiscriminativepowerthanthosefromtheoriginalstudy.
AfterpathwayanalysisontheselectedgenesbyBMI,wehavebeenabletocorrelatetheselectedgeneswithwell-knowncancer-relatedpathways.
Conclusions:OurresultsshowthatBMIcanbeusedtoanalyzemicroarraydataandtofindusefulgenesforclassifyingsamples.
PathwayanalysissuggeststhatBMIissuccessfulinidentifyingbiomarker-qualitycancer-relatedgenesfromthedata.
BackgroundLungcanceraccountsforlargeportionofcancerdeaths(29%)intheUnitedStatesformenaswellaswoman[1].
Themajortypesoflungcanceraresmall-cellandnon-small-cellcancer.
Non-small-cellcancercanbefurtherdividedintothreehistologicalsubtypes:squa-mous-cellcarcinoma,adenocarcinomaandlargecelllungcancer[2].
Regardlessofsubtype,the5-yearsurvi-valrateforlungcancerisamongthelowestofallcan-cersat15%(dataforUSA)[1].
Sincethetreatmentoflungcancerdependsonthesubtypeandthestageofcancer,itisimportanttohavedeterminedspecificmole-cularbiomarkersthatcanidentifythetypeofcancerasafunctionofgenescloselyrelatedtoeachdistinctphenotype.
Withadvanceofmicroarraytechnologies,itispossibletoconducthighthroughputdeterminationoftherela-tiverateswithwhichgenesareexpressedinagivencellortissuetype.
Thiscanhelpresearchersbetterunder-standadiseaseatthegenomiclevelandhasbecomeanimportanttoolinbiologicalsciencesaswellasmedicalandpharmaceuticalresearch.
Inthecontextoflungcan-cer,microarraytechnologycanbeusedtoidentifygeneswhoseexpressionprofileinatypeofcancerdiffersfromnormaltissuesorfromothertypesofcancer.
Suchbiomarkersareimportantsincetheycanprovidethebasisforimprovingadiagnosticclassifierorforenhan-cingthepredictionofpatient-specificprognosisor*Correspondence:glushington@ku.
edu;mvisvanathan@ku.
eduBioinformaticsCoreFacility,UniversityofKansas,Lawrence,KS66046,USALeeetal.
JournalofClinicalBioinformatics2011,1:11http://www.
jclinbioinformatics.
com/content/1/1/11JOURNALOFCLINICALBIOINFORMATICS2011Leeetal;licenseeBioMedCentralLtd.
ThisisanOpenAccessarticledistributedunderthetermsoftheCreativeCommonsAttributionLicense(http://creativecommons.
org/licenses/by/2.
0),whichpermitsunrestricteduse,distribution,andreproductioninanymedium,providedtheoriginalworkisproperlycited.
therapeuticresponse[3].
Fromaninformaticsperspec-tive,theprocessofselectingdifferentiallyexpressedgenesisreadilyachievedviadata-miningtechniquesknownasfeatureselection.
Featureselection,animpor-tantstepinthedata-miningprocess,aimstofindrepre-sentativefeaturesubsetsthatmeetdesiredcriteria.
Inmicroarraydataanalysis,onecriterionforadesiredfea-turesubsetwouldbeasetofgeneswhoseexpressionpatternsvarysignificantlywhencomparedacrossdiffer-entsamplegroups.
Theresultingsubsetcanthenbeusedtofurtheranalysissuchasbuildingadiagnosticclassifier.
Featureselectionmethods,ingeneral,canbecategor-izedintothreetypes,dependingonhowtheyarecom-binedwithotheranalysissteps:filtermethods,wrappermethodsandembeddedmethods[4].
Filtermethodsassesstherelevanceoffeaturesasscoresbylookingonlyatthepropertiesofthedata.
Featurescanbesortedbytheirscoresandlow-scoringfeaturescanberemoved.
Wrappermethodsembedtheanalysismodelwithinthefeaturesubsetsearch.
Inthissetup,asubsetoffeaturesisevaluatedbyapplyingaspecificanalysismodeltoreduceddatawiththeselectedfeaturesubset.
Inembeddedmethods,thesearchforanoptimalfeaturesubsetisbuiltintotheanalysisalgorithm.
Filtermethodsarethemostcommonlyappliedinbioinformaticsstu-diessincetheyarecomputationallysimple,fastandindependentofotheranalysisalgorithms.
Alsotheyallowfeaturestobequantifiedandprioritizedaccordingtothescores,whichisparticularlyimportantforbiologi-calinterpretation.
Inthispaper,afilter-basedfeatureselectionmethod,biomarkeridentifier(BMI),isadoptedtoanalyzegeneexpressiondatathatmightbeusedtodiscriminatebetweensampleswithandwithoutlungcancer.
Thedataconsistsofgeneexpressionpatternsinhistologi-callynormallarge-airwayepithelialcellsobtainedviabronchoscopyfromsmokers.
Genesidentifiedusingthisdatasetcanbeusedtodiagnosinglungcanceramongsmokerswithsuspectedlungcancer.
ThegenesselectedbyBMIwerecomparedwiththosefromvariousotherfeatureselectionalgorithmsandthoseidentifiedfromtheoriginalexperimentalstudy.
PathwayanalysisforthegenesselectedbyBMIwasalsoperformed.
MethodsBiomarkerIdentifierThebiomarkeridentifier(BMI)[5,6]methodcombinesvariousstatisticalmeasurestodiscerntheabilityoffea-turestodistinguishbetweentwodatagroupsofinterest.
Itconsidersthreemeasuresforevaluatingfeatures.
First,itcheckswhetherdistributionofafeatureissignificantlydifferentbetweendatagroups.
Ifthedistributionofafeaturechangessubstantially,thefeaturemightberelevanttotheunderlyingdifferencebetweendatagroups.
Second,theratioofoverallvariancerelativetovarianceincontrolgroupisusedtomeasuretherelia-bilityofafeature.
Forexample,iftheoverallvarianceisgreaterthanthatofcontrolgroup,itmeansthatthefea-turedisplaysmorenoisybehaviorinexperimentgroupmakingitlessusefulunlessitalsodemonstratesasignif-icantchangebetweendatagroup.
Ontheotherhand,anoverallvariancesmallerthanthatofcontrolgroupimpliesthatthefeatureshowsmoreconsistentbehaviorintheexperimentgroup,makingitamoreusefulfea-tureprovidedthatthereexistsasignificantdifferencebetweenthecontrasteddatagroups.
Forthesereasons,BMIpenalizesorcreditsascoreofafeaturebytheratioofoverallvariancerelativetovarianceincontrolgroup.
Lastly,BMIconsidersthediscriminativepowerofeachindividualfeaturebyincorporatingthetruepositiveratefromlogisticregressionusingthefeature.
Inmathemati-calterms,letusassumeadatasetDconsistingoftwogroups'control(ctr)'and'experiment(exp)'.
BMIassignsascoreforafeaturexdefinedasfollows:BMI(x)=λ·TP2|diff|CVctrCV,wherediff=,if≥11,otherwise.
Here,lisascalingfactorandTP2istheproductofthetruepositive(TP)ratesdeterminedforeachgroupsusinglogisticregressionoftheform'outcome~feature'.
CVctrandCVdenotethecoefficientofvarianceforthefeaturexinthe'control'groupandinbothgroups,respectively.
Also,Δ=x/xctr,wherexctr,andxdenotethemeanvalueofxin'control'andinbothgroups,respectively.
Forbiologicaldatasuchasmicroarray,thesignofΔdiffforaparticulargenecanbeinterpretedasover-expressionorunder-expressionin'experiment'comparedto'control';positiveasover-expressionandnegativeasunder-expression.
BMIhasshownpromisingresultsonvariousdatasetssuchasmassspectrometrydataofmetabolites[5],liverdisease[7]andmicroarraydatafromvarioustypesofcancer[6].
Inthisstudy,itisusedtoidentifypotentialbiomarkersforlungcancerfrommicroarraydata.
OtherfeatureselectionmethodsForcomparisonwithBMI,weused6differentpopularfeatureselectionmethods:informationgain(IG),Relief-F(RF),t-test(T)anditstwovariants(moder-atedt-test(MT)andwindowt-test(WT)),andchi-squaredtest(CS).
Leeetal.
JournalofClinicalBioinformatics2011,1:11http://www.
jclinbioinformatics.
com/content/1/1/11Page2of8InformationgainInformationgain(relativeentropy,orKullback-Leiblerdivergence),inprobabilitytheoryandinformationthe-ory,isameasureofthedifferencebetweentwoprob-abilitydistributions.
Itevaluatesafeaturexbymeasuringtheamountofinformationgainedwithrespecttotheclass(orgroup)variabley,definedasfol-lows:I(x)=H(P(y))H(P(y|x)).
Specifically,itmeasuresthedifferencebetweenthemarginaldistributionofobservableyassumingthatitisindependentoffeaturex(P(y))andtheconditionaldis-tributionofyassumingthatitisdependentofx(P(y|x)).
Ifxisnotdifferentiallyexpressed,ywillbeindepen-dentofx,thusxwillhavesmallinformationgainvalue,andviceversa.
Relief-FRelief-F[8]isaninstance-basedfeatureselectionmethodwhichevaluatesafeaturebyhowwellitsvaluedistinguishessamplesthatarefromdifferentgroupsbutaresimilartoeachother.
Foreachfeaturex,Relief-Fselectsarandomsampleandkofitsnearestneighborsfromthesameclassandeachofdifferentclasses.
Thenxisscoredasthesumofweighteddifferencesindiffer-entclassesandthesameclass.
Ifxisdifferentiallyexpressed,itwillshowgreaterdifferencesforsamplesfromdifferentclasses,thusitwillreceivehigherscore(orviceversa).
t-testandvariantsTheStudent'st-test[9]istraditionallyusedtocomparetwonormallydistributedsamplesorpopulations.
Itpre-fersfeatureswithamaximaldifferenceofmeanvaluebetweengroupsandaminimalvariabilitywithineachgroup,butitcanfailwhentherearesmallnumberofsamplesortheestimatedvariancesarenotequalbetweengroups(heteroscedasticity):scenarioswhicharecommonforpracticaldata.
Tocopewithsuchpro-blems,Welchproposedavariantoft-testtakinghetero-scedasticityintoaccount[10].
VariousstatisticaltestsfordifferentialexpressionarebasedonthetraditionalStudentandWelchtests.
Smyth[11]appliedahierarch-icalBayesianapproach(moderatedt-test)totheStudentandWelchtestsandintegratedmoreaprioriinforma-tiontoyieldmorerobustestimates.
Bergeretal.
[12]suggestedawindowt-testthatusesmultiplegeneswhichshareasimilarexpressionleveltocomputethevariancetobeincorporatedinthet-test.
Inthiswork,wechoseWelch'st-test,moderatedt-testandwindowt-testforcomparison.
chi-squaredtestChi-squaredtestisanotherpopularstatisticaltestofthedivergencebetweentheobservedandexpecteddistribu-tionofafeature.
Infeatureselection,ittestswhetherthedistributionofafeaturediffersbetweengroups.
Thechi-squarescoreusesthesummationofsquareddiffer-encesbetweenobservedandexpectedvaluesdividedbyexpectedvalues.
ExperimentaldataSpiraetal.
reportedgeneexpressiondatafromlargeair-wayepithelialcellsbymicroarrayanalysis[13].
Thisdatasetcoversasetof129AffymetrixHG-U133Amicroar-rayscomparing60smokerswithlungcancerand69smokerswithoutlungcancer.
Thisexperimentwasdesignedtodetermineifgeneexpressioninhistologi-callynormallarge-airwayepithelialcellsobtainedviabronchoscopyfromsmokerswithsuspectedlungcancercouldbeusedasalungcancerbiomarker.
Inthisdataset,7geneswereconfirmedtobedifferentiallyexpressedbetweencanceroussamplesandnon-cancer-oussamplesbyquantitativePCR[13].
TheRobustMul-tichipAverage(RMA)algorithm[14]wasusedforbackgroundadjustment,normalization,andprobe-levelsummarizationofthemicroarraysamples(pleaserefertosupplementarymethodsof[13]fordetailedinforma-tion).
Thedatasetcanbeaccessedfromgeneexpres-sionomnibus(GEO,http://www.
ncbi.
nlm.
nih.
gov/geo/)underaccessionnumberofGSE4115.
ThisdatasetwaschosensinceitconsistedofasignificantnumberofreplicatesandsomeofthegenesinthedatasetwereconfirmedbyquantitativePCR,whichprovidesagoodbasisforpreliminaryvalidation.
Tocontrastperformanceamongfeatureselectionmethods,wealsousedthedatasetpublishedthroughMicroArrayQualityControlprojectphaseII(MAQC-II).
Among9non-controldatasetsfromMAQC-II,thedatasetwiththemostbalancednumberofpositive/negativesamples(breastcancerdatawithestrogenreceptorstatusasclass)waschosen.
Thedatasetcon-sistsoftraining(130samples)andvalidation(100sam-ples)sets.
TheprocesseddatawasobtainedthroughGEOunderaccessionnumberGSE20194.
ResultsandDiscussionComparisonwithotherfeatureselectionmethodsFeatureselectionmethodscanbeevaluatedinvariousways.
Onepopularwayistoobservetheclassificationperformanceusingthefeaturesselectedbythemethod.
Ifafeatureselectionmethodisabletochoosetrulysig-nificantfeatures,theclassifiertrainedusingthosefea-turesshouldshowgoodperformancewithasmallnumberoffeatures.
Ifimportantfeaturesarealreadyknown,ontheotherhand,wecanevaluatefeatureselectionmethodsbyhowtheyrankthoseknownfea-tures.
SinceimportantfeatureshavenotbeenreportedfortheMAQC-IIdataset,itcanbeapproachedonlyviathefirstevaluationstrategy,buttheairwaydatasetisLeeetal.
JournalofClinicalBioinformatics2011,1:11http://www.
jclinbioinformatics.
com/content/1/1/11Page3of8amenabletobothmodesofevaluationsincesomeofgeneshavebeenexperimentallyconfirmedtobediffer-entiallyexpressed.
SinceaseparatevalidationsetisavailablewithintheMAQC-IIdata,weusedthetrainingsetforfeatureselec-tionandvalidationsetforclassification.
Thatis,featureselectionmethodsarefirstappliedtotrainingsettoobtainfeaturesubsets.
Then,foreachfeatureselectionmethod/classificationalgorithmpairing,classificationperformancesareevaluatedonthevalidationsetthrough10-foldcross-validationwithvaryingnumberoffeatures(from1to60).
AUCvalues(areaunderthecurve;apop-ularmeasureformodelcomparisoninmachinelearningresearchinterpretedastheprobabilitythat,givenaran-domlypickedpositiveexampleandnegativeexample,theclassifierwillassignahigherscoretothepositiveexam-plethantothenegativeone)havebeenusedhereintomeasureclassificationperformance.
LargerAUCvaluesimplymorepreciseclassification.
Forimplementation,weusedWeka[15],apopularmachinelearninglibrarywritteninJava,andthedefaultsettingwasusedforeachclassificationalgorithm.
Table1showsthemaximumAUCvalueachievedbyeachcombinationoffeatureselectionmethodsandclassificationalgorithmsfortheMAQC-IIdataset.
WecanseethattheclassifiersincombinationwithBMIshowperformancelevelscompar-abletootherswithrelativelysmallnumberoffeatures.
Also,thefeaturesselectedbyBMIshowstableperfor-manceregardlesstheclassificationalgorithm.
Fortheairwaydataset,weappliedasimilarten-foldcross-validationapproachaswiththeMAQC-IIdatatocompareclassificationperformanceofdifferentfeatureselectionmethods.
Here,thedatawasdividedinto10-folds,whereby9foldsareusedforbothselectingfeaturesandtrainingclassifiers,andthereservedfoldwasusedtocalculateAUCvalueoftrainedclassifiers.
Foreachcombi-nationoffeatureselectionmethodsandclassificationalgo-rithms,thisprocesswasrepeated10timeswithadifferentreservedfold,whilevaryingnumberoffeatures(from1to60)andtheAUCvalueswereaveragedoverthetendis-tinctreserved-foldcases.
TheparametersettingforeachclassificationalgorithmwasthesameasinMAQC-IIdataset.
Table2showsthemaximumAUCvalueachievedbyeachcombinationoffeatureselectionmethodsandclassi-ficationalgorithms.
AsinMAQC-IIdataset,theclassifiersincombinationwithBMIshowcomparableperformancewithotherswithrelativelysmallnumberoffeatures.
AndthefeaturesselectedbyBMIshowstableperformanceregardlesstheclassificationalgorithm.
Next,fortheairwaydataset,weinvestigatedhowthegenesconfirmedintheliterature(DUOX1,BACH2,DCLRE1C,RAB1A,TPD52,FOS,andIL8)arerankedbyBMIcomparedtootherfeatureselectionmethods.
Ifthesegenesaregenerallyrankedhighly,afeatureTable1ComparisonofclassificationperformancesonMAQC-IIdatasetClassificationAlgorithmsFeatureSelectionMethodsSupportVectorMachinek-NearestNeighborNaiveBayesRandomForestInformationGain0.
9031(6)0.
9380(25)0.
9008(40)0.
9206(50)Chi-squaredtest0.
8821(1)0.
9164(50)0.
9151(4)0.
9441(60)Relief-F0.
8821(1)0.
9052(15)0.
8995(50)0.
9306(60)t-test0.
9067(15)0.
9100(20)0.
9042(8)0.
9304(40)Windowt-test0.
8903(5)0.
9216(5)0.
9012(2)0.
9199(10)Moderatedt-test0.
8903(6)0.
9084(5)0.
8987(1)0.
9309(50)BMI0.
9077(4)0.
9298(15)0.
9164(4)0.
9250(9)EachvaluerepresentsthemaximumAUCvalue(by10-foldcross-validation)achievedbythecorrespondingfeatureselectionmethodandclassificationalgorithm.
Thenumberoffeaturesusedtoachievethemaximumisshowninsideparenthesis.
Table2ComparisonofclassificationperformancesonairwaydatasetClassificationAlgorithmsFeatureSelectionMethodsSupportVectorMachinek-NearestNeighborNaiveBayesRandomForestInformationGain0.
6853(40)0.
8006(4)0.
8297(50)0.
8620(60)Chi-squaredtest0.
7052(20)0.
8029(60)0.
7997(3)0.
8309(50)Relief-F0.
6633(25)0.
7825(9)0.
8329(25)0.
8685(60)t-test0.
6902(8)0.
7822(4)0.
8402(4)0.
8121(8)Windowt-test0.
6856(20)0.
7817(30)0.
8367(20)0.
8093(40)Moderatedt-test0.
6878(6)0.
7875(5)0.
8329(5)0.
8115(20)BMI0.
7572(9)0.
8005(5)0.
8299(5)0.
8212(10)EachvaluerepresentsthemaximumAUCvalue(via10-foldcross-validation)achievedbythecorrespondingfeatureselectionmethodandclassificationalgorithm.
Thenumberoffeaturesusedtoachievethemaximumisshowninsideparenthesis.
Leeetal.
JournalofClinicalBioinformatics2011,1:11http://www.
jclinbioinformatics.
com/content/1/1/11Page4of8selectionmethodcouldbesaidtocorroboratethegivendata.
Asbefore,wedividedthedatainto10foldsandusedonly9foldsinfeatureselection,repeatingthefea-tureselectionforeachdistinctreservedfold.
Foreachofthesetenfoldcases,werecordedgeneranksasdeter-minedbyeachmethodandcalculatedthemedianvalueforeachgene.
Figure1showsmedianranksofvalidatedgenesbydifferentfeatureselectionmethods,demonstrat-ingthatBMIranksalloftheconfirmedgeneswithinthetop4000rankedgenes,andtheoverallBMIrankingofconfirmedgenesisgenerallysuperiortoothermethods.
Fromtheseresults,itcanbesaidthatBMIshowscompetitiveperformanceinidentifyingusefulfeaturesforclassificationandshowshighconsistencywithactualdifferentialexpression.
ComparisonwithbiomarkersfromliteratureFortheairwaydataset,wefurthercomparedthegenesselectedbyBMIandthebiomarkersfromoriginalliterature[13].
Inoriginalliterature,80featureswereselectedtodistinguishcanceroussamplesfromnormalsamples.
ForBMI,wechose10featuresthatwereusedtoachievethebestclassificationperformanceinTable2.
Theselected10fea-turesareshowninTable3.
Thenwetrainedvariouspopularclassificationalgorithmsusingthesetwosetsoffeatures:naveBayes,supportvectormachine(SVM),neuralnetwork,k-nearestneighbor,andrandomforest.
Weusedtheimple-mentationinWekasoftware[15]withdefaultsettings.
Table4showsthedetailedclassificationperformancesobtainedfrom20independentrunsof10-foldcross-validation.
ClassifierstrainedusingfeaturesselectedbyBMIgenerallyshowedbetterperformanceformostclas-sificationalgorithms.
ThisimpliesthatthefeaturesselectedbyBMIaremoreusefulforconstructingaccu-rateclassifiers,whichcanprovideagoodbasisforfurtherscreeningofbiomarkers.
PathwayanalysisofselectedbiomarkersAlthoughasetofgenesisusefulfortrainingclassifier,theconstituentgenesmaybeuselessasbiomarkersifFigure1Themedianranksofvalidatedgenesinairwaydatasetbyvariousfeatureselectionmethods.
Table3Top10genesselectedbyBMIProbeIDSymbolRegulationName201694_s_atEGR1Upearlygrowthresponse1202056_atKPNA1Upkaryopherinalpha1(importinalpha5)203265_s_atMAP2K4Upmitogen-activatedproteinkinasekinase4207283_atRPL23AP13DownribosomalproteinL23apseudogene13211612_s_atIL13RA1Upinterleukin13receptor,alpha1214261_s_atADH6Upalcoholdehydrogenase6(classV)216609_atTXNDownFulllengthinsertcDNAcloneYI46D09219233_s_atGSDMBDowngasderminB222339_x_at-Down-34206atARAP1DownArfGAPwithRhoGAPdomain,ankyrinrepeatandPHdomain1Leeetal.
JournalofClinicalBioinformatics2011,1:11http://www.
jclinbioinformatics.
com/content/1/1/11Page5of8theirbiologicalrolesarenotrelatedtothetargetdiseaseorprocess.
Thusweanalyzedthepathwaysassociatedwith80highly-rankedgenestoinvestigatetheirbiologi-calroles.
Forpathwayanalysis,weinvestigatedasso-ciatedtermsinKEGGpathways[16],NCI-Naturepathwayinteractiondatabase[17],andPANTHER(pro-teinanalysisthroughevolutionaryrelationships)classifi-cationsystem[18]usingtheEGANprogram[19].
Tables5and6summarizethegenesandtheirasso-ciatedpathwayswithsignificantp-values(<0.
05).
Wecanobservethattherearesomegenes(EGR1,FOS,DUSP10,andMAP2K4)associatedwithmitogen-acti-vatedproteinkinase(MAPK)pathways,whichisawell-knowntargetintheoncologydrugdiscovery[20].
Also,threegenes(APC,MSH2,andATF3)showedsignificantassociationwithatermfromtheNCI-NaturePathwayInteractionDatabase,'Directp53effectors.
'Thisimpliesthatthosegenesarerelatedwithprotein'p53'whichisknownasatumorsuppressorprotein[21].
WenotethatincidenceofthegeneralKEGGannotation'path-waysincancer'showedagoodassociation(p-valueof0.
0019)withoursetof80genes.
Onealsofindsotherpathwaysrelatedwithknownoncogenessuchasc-Met[22]andepidermalgrowthfactorreceptor(EGFRorErbB-1)[23]withinourlist.
Fromthese,itcanbesaidthatgeneshighlyrankedbyBMIaregenerallyrelevanttocancerdevelopmentordiagnosis,thusBMIappearstobeusefulforidentifyingpotentialbiomarkersforlungcancer.
ConclusionsInthiswork,afilter-basedfeatureselectionmethod,biomarkeridentifier(BMI),hasbeenappliedtofindpotentialbiomarkersforlungcancerfrommicroarrayTable4ClassificationperformanceswithselectedbiomarkersbyBMIandoriginalliteratureBiomarkersbyBMIBiomarkersfromoriginalliteratureClassifierSpecificitySensitivityAccuracySpecificitySensitivityAccuracyNaveBayes0.
7938++0.
7006++0.
7489++0.
71170.
66440.
6872SVM0.
8134++0.
7056++0.
7615++0.
66220.
65930.
6607NeuralNetwork0.
7242++0.
64220.
68480.
69560.
7459++0.
7217++k-NearestNeighbor0.
8325++0.
61440.
7275++0.
63780.
6964++0.
6682RandomForest0.
7139++0.
7328++0.
7230++0.
68720.
66800.
6773++and+denotessuperiorperformanceasdeterminedatof1%and5%significancelevelsrespectively.
Table5KEGGpathwaysandPANTHERclassificationsassociatedwithtop80genesselectedbyBMIKEGGpathwaynamep-valueAssociatedgenesColorectalcancer1.
3809E-4FOS,MSH2,APCPathwaysincancer0.
0019FOS,MSH2,APC,TCEB2Metabolicpathways0.
0021ADH6,SAT1,EXT2,TGDS,BTD,PRPS1,AGPSBiotinmetabolism0.
0032BTDMAPKsignalingpathway0.
0094DUSP10,MAP2K4,FOSCytokine-cytokinereceptorinteraction0.
0098CXCR4,ACVR2A,IL13RA1Toll-likereceptorsignalingpathway0.
0117FOS,MAP2K4Tightjunction0.
0196PPP2R2D,INADLMismatchrepair0.
0361MSH2Glycosaminoglycanbiosynthesis-heparansulfate0.
0408EXT2Pentosephosphatepathway0.
0423PRPS1Endocytosis0.
0428ARAP1,CXCR4PANTHERclassificationp-valueAssociatedgenesOxidativestressresponse8.
6417E-5TXN,MAP2K4,DUSP10O-antigenbiosynthesis0.
0064TGDSTcellactivation0.
0083FOS,B2MInterleukinsignalingpathway0.
0108IL13RA1,FOSApoptosissignalingpathway0.
0133ATF3,FOSFGFsignalingpathway0.
0135MAP2K4,PPP2R2DAxonguidancemediatedbySlit/Robo0.
0253CXCR4HypoxiaresponseviaHIFactivation0.
0408TXNInsulin/IGFpathway-mitogenactivatedproteinkinasekinase/MAPkinasecascade0.
0484FOSLeeetal.
JournalofClinicalBioinformatics2011,1:11http://www.
jclinbioinformatics.
com/content/1/1/11Page6of8data.
BMImeasuresthepotentialvalueofeachgeneasabiomarkercandidatebycombiningvariousstatisticalmeasurestoassessitsabilitytodistinguishbetweentwodatagroupsofinterest.
WeevaluatedBMIperformanceontwopublicmicroarraydatasets:onefromtheMicro-ArrayQualityControlprojectandtheotherfromsmo-kerswithandwithoutlungcancer.
BMIwascomparedwithotherpopularfilter-basedfeatureselectionmethodsonbothdatasetandshowedcompetitiveperformanceinselectingusefulfeaturesforvariousclassificationalgo-rithms.
SinceofthelatterdatasetincludesinformationregardingspecificgeneswhosetissuedifferentiationrelevancehasbeenvalidatedbyquantitativeRT-PCR,wealsocomparedhowthesegeneswererankedbydif-ferentfeatureselectionalgorithm.
ThevalidatedgenesgenerallywereassignedhigherranksbyBMIthanbyothermethods,implyingthatBMIshouldbeeffectiveinidentifyingbiomarkersthatshowdifferentialexpressionincanceroussamples.
WealsocomparedBMIwiththeapproachintheoriginalanalysisconductedonthelungcancermicroarraydata[13]bycontrastingtheclassifica-tionperformanceusingselectedgenesfromeachTable6NCI-Naturepathwayinteractionsassociatedwithtop80genesselectedbyBMINCI-NaturePathwayInteractionp-valueAssociatedgenesATF-2transcriptionfactornetwork6.
8276E-5ATF3,FOS,DUSP10DownstreamsignalinginnaveCD8+Tcells1.
8173E-4B2M,EGR1,FOSSignalingeventsmediatedbyHepatocyteGrowthFactorReceptor(c-Met)2.
6255E-4EGR1,MAP2K4,APCEphrinBreversesignaling8.
6116E-4CXCR4,MAP2K4ErbB1downstreamsignaling8.
7013E-4MAP2K4,FOS,EGR1Regulationofp38-alphaandp38-beta0.
0011DUSP10,MAP2K4Directp53effectors0.
0013APC,MSH2,ATF3TrkreceptorsignalingmediatedbytheMAPKpathway0.
0014EGR1,FOSRhoAsignalingpathway0.
0021FOS,MAP2K4IL6-mediatedsignalingevents0.
0023MAP2K4,FOSPresenilinactioninNotchandWntsignaling0.
0024FOS,APCCalcineurin-regulatedNFAT-dependenttranscriptioninlymphocytes0.
0025EGR1,FOSRegulationofAndrogenreceptoractivity0.
0027EGR1,MAP2K4Fc-epsilonreceptorIsignalinginmastcells0.
0041FOS,MAP2K4IL12-mediatedsignalingevents0.
0045B2M,FOSHIF-1-alphatranscriptionfactornetwork0.
0052FOS,CXCR4CDC42signalingevents0.
0058APC,MAP2K4RegulationofnuclearSMAD2/3signaling0.
0075FOS,ATF3Glucocorticoidreceptorregulatorynetwork0.
0077FOS,EGR1SumoylationbyRanBP2regulatestranscriptionalrepression0.
0174RANBP2JNKsignalingintheCD4+TCRpathway0.
0206MAP2K4RassignalingintheCD4+TCRpathway0.
0222FOSHypoxicandoxygenhomeostasisregulationofHIF-1-alpha0.
0284TCEB2CellularrolesofAnthraxtoxin0.
0346MAP2K4VEGFR3signalinginlymphaticendothelium0.
0361MAP2K4S1P2pathway0.
0377FOSPDGFR-alphasignalingpathway0.
0377FOSALK1signalingevents0.
0392ACVR2ASignalingeventsmediatedbyPRL0.
0392EGR1TRAILsignalingpathway0.
0438MAP2K4RegulationofCDC42activity0.
0453APCS1P3pathway0.
0453CXCR4CD40/CD40Lsignaling0.
0469MAP2K4CanonicalWntsignalingpathway0.
0469APCp38MAPKsignalingpathway0.
0469TXNCalciumsignalingintheCD4+TCRpathway0.
0484FOSNongenotropicAndrogensignaling0.
0484FOSNephrin/Neph1signalinginthekidneypodocyte0.
0499MAP2K4IL12signalingmediatedbySTAT40.
0499FOSLeeetal.
JournalofClinicalBioinformatics2011,1:11http://www.
jclinbioinformatics.
com/content/1/1/11Page7of8method.
Givenmodelstrainedforvariousclassificationalgorithms,classifiersbasedongenesselectedbyBMIshowedbetterperformancethanthosefromoriginalstudy.
Finally,inevaluatingwhetherthegenesselectedbyBMIhaveknownbiologicalfunctionrelatedto(lung)cancer,weanalyzedtheirpathwaydispositionandfoundthatmanygeneswereassociatedwithknowncancer-relatedpathways.
ThuswecanconcludethatBMIisasuitabletechniqueforphenotypicclassificationofmicro-arraydataandmayprovideareasonablemechanismforidentifyingviablediagnosticbiomarkercandidates.
Basedontheresultsinthisstudy,wearepursuingafol-low-upstudyusingBMItoidentifybiomarkerssuitableforthelungcanceranalysiswithexperimentaldataonclinicallyderivedtissues.
AcknowledgementsThispublicationwasmadepossiblebygrantnumberP20RR016475fromtheNationalCenterforResearchResources(NCRR),acomponentoftheNationalInstitutesofHealth(NIH).
WealsowouldliketothankDrs.
MichaelNetzerandChristianBaumgartnerfromUniversityofHealthSciences,MedicalInformaticsandTechnology(UMIT),AustriainprovidingsourcecodeforBMIimplementation.
Authors'contributionsILparticipatedinthedesignofthestudy,performedthestatisticalanalysisanddraftedthemanuscript.
GLandMVconceivedofthestudy,andparticipatedinitsdesignandcoordination.
Allauthorsreadandapprovedthefinalmanuscript.
CompetinginterestsTheauthorsdeclarethattheyhavenocompetinginterests.
Received:8October2010Accepted:21March2011Published:21March2011References1.
JemalA,SiegelR,WardE,HaoY,XuJ,MurrayT,ThunMJ:Cancerstatistics.
CACancerJClin2008,58:71-96.
2.
HerbstRS,HeymachJV,LippmanSM:Lungcancer.
NewEnglandJournalofMedicine2008,359:1367-1380.
3.
GranvilleCA,DennisPA:Anoverviewoflungcancergenomicsandproteomics.
AmericanJournalofRespiratoryCellandMolecularBiology2005,32:169-176.
4.
SaeysY,InzaI,LarraagaP:Areviewoffeatureselectiontechniquesinbioinformatics.
Bioinformatics2007,23:2507-2517.
5.
BaumgartnerC,BaumgartnerD:Biomarkerdiscovery,diseaseclassification,andsimilarityqueryprocessingonhigh-throughputMS/MSdataofinbornerrorsofmetabolism.
JournalofBiomolecularScreening2006,11:90-99.
6.
VisvanathanM,NetzerM,SegerM,AdagarlaBS,BaumgartnerC,SittampalamS,LushingtonGH:Oncogenesandpathwayidentificationusingfilter-basedapproachesbetweenvariouscarcinomatypesinlung.
InternationalJournalofComputationalBiologyandDrugDesign2009,2:236-251.
7.
NetzerM,MillonigG,OslM,PfeiferB,PraunS,VillingerJ,VogelW,BaumgartnerC:Anewensemble-basedalgorithmforidentifyingbreathgasmarkercandidatesinliverdiseaseusingionmoleculereactionmassspectrometry.
Bioinformatics2009,25(7):941-947.
8.
KononenkoI:Estimatingattributes:analysisandextensionsofRELIEF.
InECML-94:ProceedingsoftheEuropeanconferenceonmachinelearningonMachineLearning.
Editedby:BergadanoF,DeRaedtL.
SpringerBerlin/Heidelberg;1994:171-182.
9.
Student:Theprobableerrorofamean.
Biometrika1908,6:1-25.
10.
WelchBL:Thesignificanceofthedifferencebetweentwomeanswhenthepopulationvariancesareunequal.
Biometrika1938,29:350-362.
11.
SmythGK:LinearmodelsandempiricalBayesmethodsforassessingdifferentialexpressioninmicroarrayexperiments.
StatisticalApplicationsinGeneticsandMolecularBiology2004,3:3.
12.
BergerF,DeHertoghB,PierreM,GaigneauxA,DepiereuxE:The"Windowt-test":asimpleandpowerfulapproachtodetectdifferentiallyexpressedgenesinmicroarraydatasets.
CentralEuropeanJournalofBiology2008,3:327-344.
13.
SpiraA,BeaneJE,ShahV,SteilingK,LiuG,SchembriF,GilmanS,DumasYM,CalnerP,SebastianiP,SridharS,BeamisJ,LambC,AndersonT,GerryN,KeaneJ,LenburgME,BrodyJS:Airwayepithelialgeneexpressioninthediagnosticevaluationofsmokerswithsuspectlungcancer.
NatureMedicine2007,13:361-366.
14.
IrizarryRA,BolstadBM,CollinF,CopeLM,HobbsB,SpeedTP:SummariesofAffymetrixGeneChipprobeleveldata.
NucleicAcidsResearch2003,31:e15.
15.
HallM,FrankE,HolmesG,PfahringerB,ReutemannP,WittenIH:TheWEKADataMiningSoftware:AnUpdate.
Explorations2009,11:10-18.
16.
KanehisaM,GotoS,FurumichiM,TanabeM,HirakawaM:KEGGforrepresentationandanalysisofmolecularnetworksinvolvingdiseasesanddrugs.
NucleicAcidsResearch2010,38:D355-D360.
17.
SchaeferCF,AnthonyK,KrupaS,BuchoJ,DayM,HannayT,BuetowKH:PID:thepathwayinteractiondatabase.
NucleicAcidsResearch2009,37:D674-D679.
18.
ThomasPD,CampbellMJ,KejariwalA,MiH,KarlakB,DavermanR,DiemerK,MuruganujanA,NarechaniaA:PANTHER:alibraryofproteinfamiliesandsubfamiliesindexedbyfunction.
GenomeResearch2003,13:2129-2141.
19.
PaquetteJ,TokuyasuT:EGAN:exploratorygeneassociationnetworks.
Bioinformatics2010,26:285-286.
20.
Sebolt-LeopoldJS:AdvancesinthedevelopmentofcancertherapeuticsdirectedagainsttheRAS-mitogen-activatedproteinkinasepathway.
ClinicalCancerResearch2008,14:3651-3656.
21.
HollsteinM,SidranskyD,VogelsteinB,HarrisCC:p53mutationsinhumancancers.
Science1991,253:49-53.
22.
SattlerM,SalgiaR:c-Metandhepatocytegrowthfactor:Potentialasnoveltargetsincancertherapy.
CurrentOncologyReports2007,9:102-108.
23.
ZhangH,BerezovA,WangQ,ZhangG,DrebinJ,MuraliR,GreeneMI:ErbBreceptors:fromoncogenestotargetedcancertherapies.
TheJournalofClinicalInvestigation2007,117:2051-2058.
doi:10.
1186/2043-9113-1-11Citethisarticleas:Leeetal.
:Afilter-basedfeatureselectionapproachforidentifyingpotentialbiomarkersforlungcancer.
JournalofClinicalBioinformatics20111:11.
SubmityournextmanuscripttoBioMedCentralandtakefulladvantageof:ConvenientonlinesubmissionThoroughpeerreviewNospaceconstraintsorcolorgurechargesImmediatepublicationonacceptanceInclusioninPubMed,CAS,ScopusandGoogleScholarResearchwhichisfreelyavailableforredistributionSubmityourmanuscriptatwww.
biomedcentral.
com/submitLeeetal.
JournalofClinicalBioinformatics2011,1:11http://www.
jclinbioinformatics.
com/content/1/1/11Page8of8
- Smythwww.07ppp.com相关文档
- decreasingwww.07ppp.com
- contextwww.07ppp.com
- indicationwww.07ppp.com
- processedwww.07ppp.com
- Dealswww.07ppp.com
- exceedswww.07ppp.com
香港CN2云服务器 1核 2G 35元/月 妮妮云
妮妮云的来历妮妮云是 789 陈总 张总 三方共同投资建立的网站 本着“良心 便宜 稳定”的初衷 为小白用户避免被坑妮妮云的市场定位妮妮云主要代理市场稳定速度的云服务器产品,避免新手购买云服务器的时候众多商家不知道如何选择,妮妮云就帮你选择好了产品,无需承担购买风险,不用担心出现被跑路 被诈骗的情况。妮妮云的售后保证妮妮云退款 通过于合作商的友好协商,云服务器提供2天内全额退款到网站余额,超过2天...
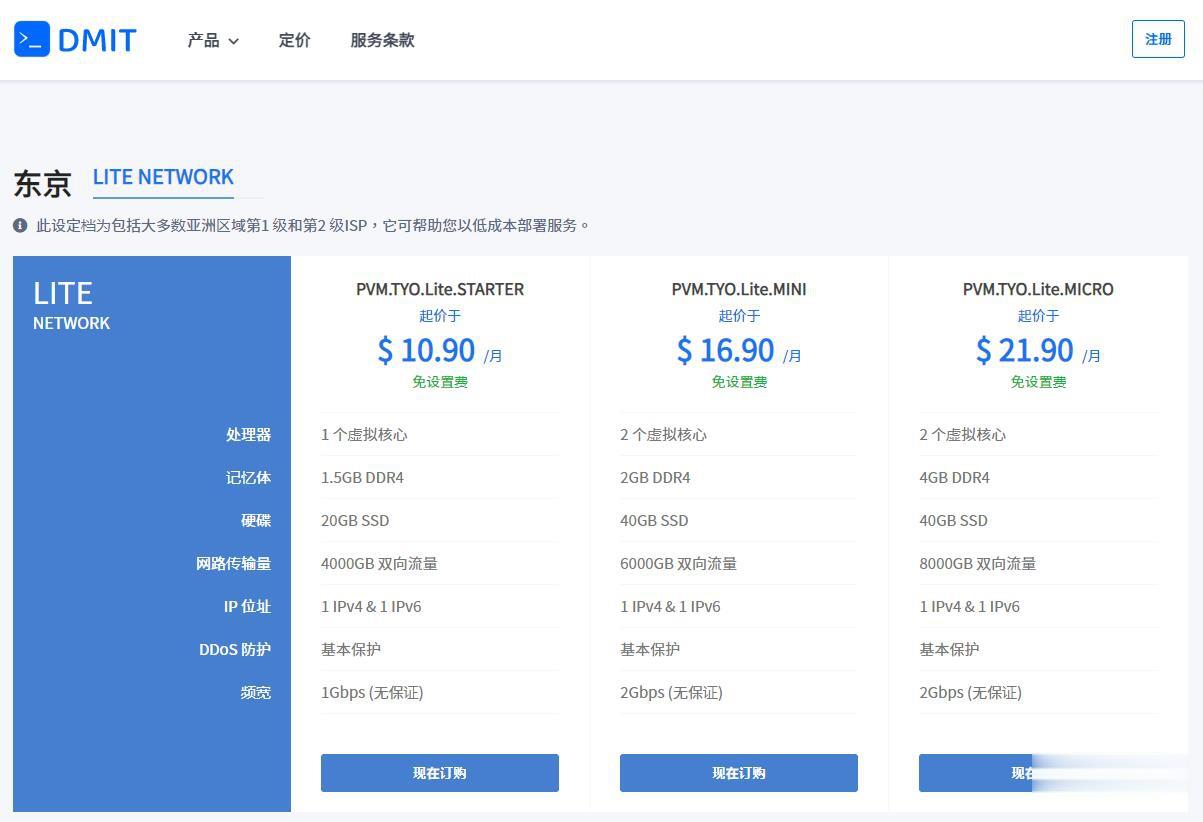
DMIT$10.9/月,日本VPS/三网直连/1核1.5G内存/20GB存储/1Gbps端口
优惠码年付一次性5折优惠码:TYO-Lite-Open-Beta-1y-50OFF永久8折优惠码:TYO-Lite-Open-Beta-Recur-20OFF日本vpsCPU内存SSD流量带宽价格购买1核1.5G20 GB4 TB1Gbps$10.9/月购买2核2 G40 GB6 TB1Gbps$16.9/月购买2核4 G60 GB8 TB1Gbps$21.9/月购买4核4 G80 GB12 TB...
百星数据(60元/月,600元/年)日本/韩国/香港cn2 gia云服务器,2核2G/40G/5M带宽
百星数据(baixidc),2012年开始运作至今,主要提供境外自营云服务器和独立服务器出租业务,根据网络线路的不同划分为:美国cera 9929、美国cn2 gia、香港cn2 gia、韩国cn2 gia、日本cn2 gia等云服务器及物理服务器业务。目前,百星数据 推出的日本、韩国、香港cn2 gia云服务器,2核2G/40G/5M带宽低至60元/月,600元/年。百星数据优惠码:优惠码:30...

www.07ppp.com为你推荐
-
百度商城百度商城知道在哪个地方,怎么找不到啊甲骨文不满赔偿不签合同不满一年怎么补偿巫正刚想在淘宝开一个类似于耐克、阿迪之类的店、需要多少钱、如何能够代理同ip站点同IP网站具体是什么意思,能换独立的吗长尾关键词挖掘工具外贸长尾关键词挖掘工具哪个好用sss17.comwww.com17com.com是什么啊?www.gegeshe.com有什么好听的流行歌曲125xx.com115xx.com是什么意思yinrentangzimotang氨基酸洗发水的功效咋样?dadi.tvApple TV是干嘛的?怎么用?多少钱?